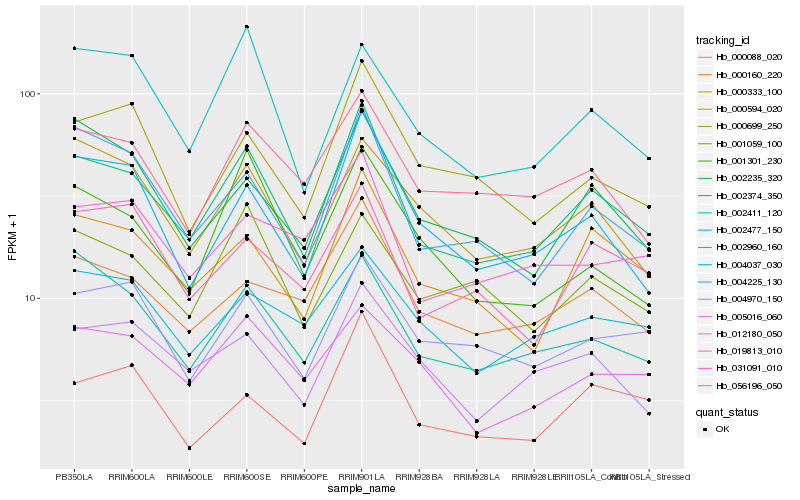
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002374_350 |
0.0 |
- |
- |
PREDICTED: delta-aminolevulinic acid dehydratase, chloroplastic [Populus euphratica] |
| 2 |
Hb_000333_100 |
0.0862452599 |
- |
- |
PREDICTED: tubulin--tyrosine ligase-like protein 12 isoform X1 [Jatropha curcas] |
| 3 |
Hb_004225_130 |
0.0938197265 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105628060 isoform X1 [Jatropha curcas] |
| 4 |
Hb_004037_030 |
0.0991863619 |
- |
- |
PREDICTED: DNA repair endonuclease UVH1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000160_220 |
0.1008378046 |
- |
- |
PREDICTED: protein AAR2 homolog isoform X1 [Jatropha curcas] |
| 6 |
Hb_002411_120 |
0.1031279007 |
- |
- |
PREDICTED: uncharacterized protein LOC105631645 [Jatropha curcas] |
| 7 |
Hb_056196_050 |
0.1040719676 |
- |
- |
PREDICTED: nuclear poly(A) polymerase 4-like [Jatropha curcas] |
| 8 |
Hb_004970_150 |
0.105280973 |
- |
- |
PREDICTED: uncharacterized protein LOC105633636 [Jatropha curcas] |
| 9 |
Hb_005016_060 |
0.1061549311 |
- |
- |
PREDICTED: histidine--tRNA ligase, cytoplasmic [Jatropha curcas] |
| 10 |
Hb_002235_320 |
0.1100892583 |
- |
- |
PREDICTED: polypyrimidine tract-binding protein homolog 1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_031091_010 |
0.1122995961 |
- |
- |
crooked neck protein, putative [Ricinus communis] |
| 12 |
Hb_000594_020 |
0.1124052989 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 25 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000699_250 |
0.11248174 |
- |
- |
Poly(rC)-binding protein, putative [Ricinus communis] |
| 14 |
Hb_019813_010 |
0.1133110891 |
- |
- |
PREDICTED: uncharacterized protein LOC105646947 [Jatropha curcas] |
| 15 |
Hb_012180_050 |
0.1137991115 |
- |
- |
PREDICTED: uncharacterized protein LOC105632355 isoform X3 [Jatropha curcas] |
| 16 |
Hb_001059_100 |
0.1143776317 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein U-like protein 1 [Jatropha curcas] |
| 17 |
Hb_000088_020 |
0.1146278423 |
transcription factor |
TF Family: M-type |
hypothetical protein JCGZ_10946 [Jatropha curcas] |
| 18 |
Hb_002960_160 |
0.1157757097 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 35B isoform X2 [Jatropha curcas] |
| 19 |
Hb_002477_150 |
0.1158282851 |
transcription factor |
TF Family: Alfin-like |
DNA binding protein, putative [Ricinus communis] |
| 20 |
Hb_001301_230 |
0.1167713141 |
- |
- |
PREDICTED: uncharacterized protein LOC105632949 isoform X2 [Jatropha curcas] |