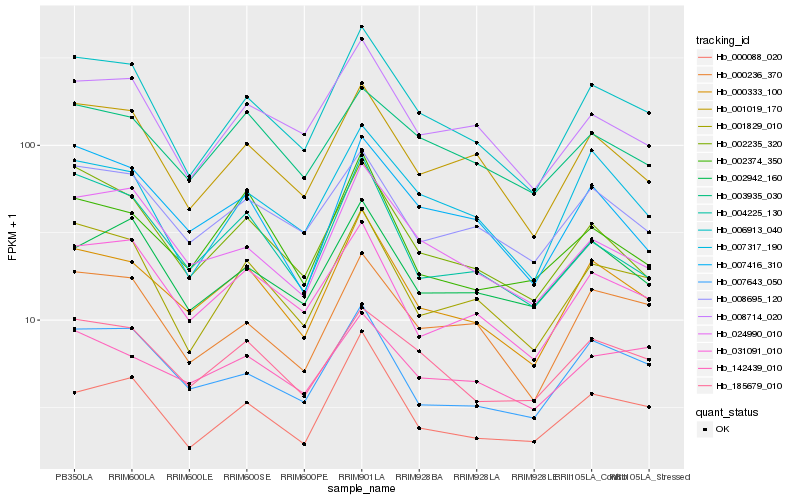
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000333_100 |
0.0 |
- |
- |
PREDICTED: tubulin--tyrosine ligase-like protein 12 isoform X1 [Jatropha curcas] |
| 2 |
Hb_006913_040 |
0.0710950138 |
- |
- |
PREDICTED: hydroxyacylglutathione hydrolase cytoplasmic [Jatropha curcas] |
| 3 |
Hb_007643_050 |
0.0838172005 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_031091_010 |
0.083981113 |
- |
- |
crooked neck protein, putative [Ricinus communis] |
| 5 |
Hb_008695_120 |
0.0857558939 |
transcription factor |
TF Family: C2H2 |
PREDICTED: MYST-like histone acetyltransferase 2 [Jatropha curcas] |
| 6 |
Hb_002374_350 |
0.0862452599 |
- |
- |
PREDICTED: delta-aminolevulinic acid dehydratase, chloroplastic [Populus euphratica] |
| 7 |
Hb_001019_170 |
0.0868998425 |
- |
- |
PREDICTED: uncharacterized protein LOC105639322 [Jatropha curcas] |
| 8 |
Hb_185679_010 |
0.0898744436 |
- |
- |
serine/threonine-protein kinase rio2, putative [Ricinus communis] |
| 9 |
Hb_001829_010 |
0.09104901 |
- |
- |
PREDICTED: general transcription factor 3C polypeptide 3-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_002235_320 |
0.0927095723 |
- |
- |
PREDICTED: polypyrimidine tract-binding protein homolog 1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_007416_310 |
0.0953583478 |
- |
- |
PREDICTED: zinc finger protein 622 [Jatropha curcas] |
| 12 |
Hb_000088_020 |
0.0957749381 |
transcription factor |
TF Family: M-type |
hypothetical protein JCGZ_10946 [Jatropha curcas] |
| 13 |
Hb_007317_190 |
0.096215998 |
- |
- |
PREDICTED: WD-40 repeat-containing protein MSI1 [Jatropha curcas] |
| 14 |
Hb_008714_020 |
0.0971179592 |
- |
- |
PREDICTED: GTP-binding protein SAR1A-like [Jatropha curcas] |
| 15 |
Hb_000236_370 |
0.0973306713 |
- |
- |
PREDICTED: DNA-directed RNA polymerases I and III subunit RPAC1 [Jatropha curcas] |
| 16 |
Hb_004225_130 |
0.0975613876 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105628060 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002942_160 |
0.0987082397 |
- |
- |
PREDICTED: coiled-coil domain-containing protein SCD2 [Jatropha curcas] |
| 18 |
Hb_024990_010 |
0.0994379664 |
- |
- |
PREDICTED: la protein 1 [Jatropha curcas] |
| 19 |
Hb_003935_030 |
0.1001135078 |
- |
- |
PREDICTED: 14-3-3-like protein GF14 kappa isoform X2 [Jatropha curcas] |
| 20 |
Hb_142439_010 |
0.1008312672 |
- |
- |
PREDICTED: F-box/kelch-repeat protein OR23 [Jatropha curcas] |