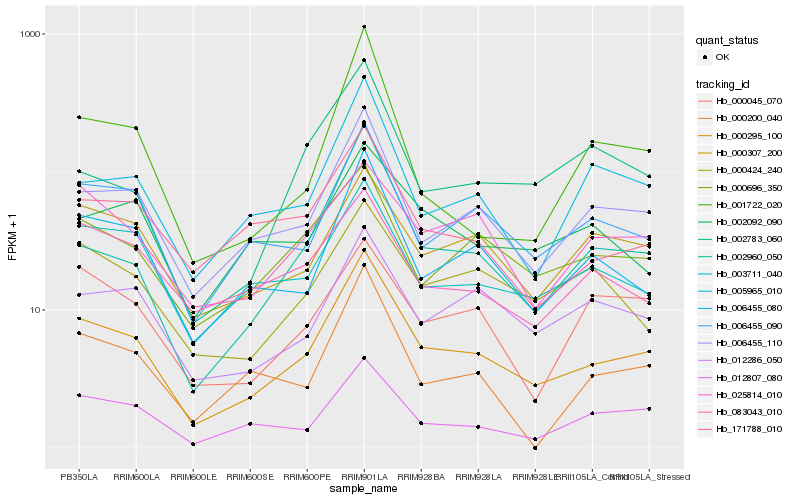
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000295_100 |
0.0 |
- |
- |
- |
| 2 |
Hb_006455_110 |
0.1071750306 |
- |
- |
bromodomain-containing protein, putative [Ricinus communis] |
| 3 |
Hb_006455_080 |
0.1204176559 |
- |
- |
PREDICTED: transcription factor GTE6-like isoform X2 [Nelumbo nucifera] |
| 4 |
Hb_006455_090 |
0.1244561151 |
- |
- |
PREDICTED: transcription factor GTE1-like [Jatropha curcas] |
| 5 |
Hb_002960_050 |
0.127874474 |
- |
- |
PREDICTED: uncharacterized protein LOC105635325 [Jatropha curcas] |
| 6 |
Hb_012807_080 |
0.1369291155 |
- |
- |
hypothetical protein CICLE_v10028099mg [Citrus clementina] |
| 7 |
Hb_005965_010 |
0.1409121654 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb family transcription factor APL-like [Jatropha curcas] |
| 8 |
Hb_000696_350 |
0.141520803 |
- |
- |
ARM repeat superfamily protein isoform 2 [Theobroma cacao] |
| 9 |
Hb_003711_040 |
0.143716523 |
- |
- |
PREDICTED: UBX domain-containing protein 4 [Jatropha curcas] |
| 10 |
Hb_012286_050 |
0.1459284933 |
- |
- |
PREDICTED: syntaxin-61 [Jatropha curcas] |
| 11 |
Hb_171788_010 |
0.1489006887 |
- |
- |
hypothetical protein CISIN_1g007892mg [Citrus sinensis] |
| 12 |
Hb_000307_200 |
0.1515549211 |
- |
- |
PREDICTED: uncharacterized protein LOC105638529 [Jatropha curcas] |
| 13 |
Hb_000424_240 |
0.1591201517 |
- |
- |
PREDICTED: 3beta-hydroxysteroid-dehydrogenase/decarboxylase isoform 2 [Jatropha curcas] |
| 14 |
Hb_000200_040 |
0.1635404685 |
- |
- |
- |
| 15 |
Hb_025814_010 |
0.165665388 |
- |
- |
hypothetical protein PRUPE_ppa015585mg [Prunus persica] |
| 16 |
Hb_002783_060 |
0.1671533059 |
- |
- |
PREDICTED: uncharacterized protein LOC105635325 [Jatropha curcas] |
| 17 |
Hb_001722_020 |
0.1679846262 |
- |
- |
- |
| 18 |
Hb_002092_090 |
0.1689968776 |
- |
- |
PREDICTED: aspartic proteinase Asp1 [Jatropha curcas] |
| 19 |
Hb_083043_010 |
0.1704928163 |
- |
- |
PREDICTED: ribonuclease P protein subunit p25-like protein [Jatropha curcas] |
| 20 |
Hb_000045_070 |
0.1711240499 |
- |
- |
conserved hypothetical protein [Ricinus communis] |