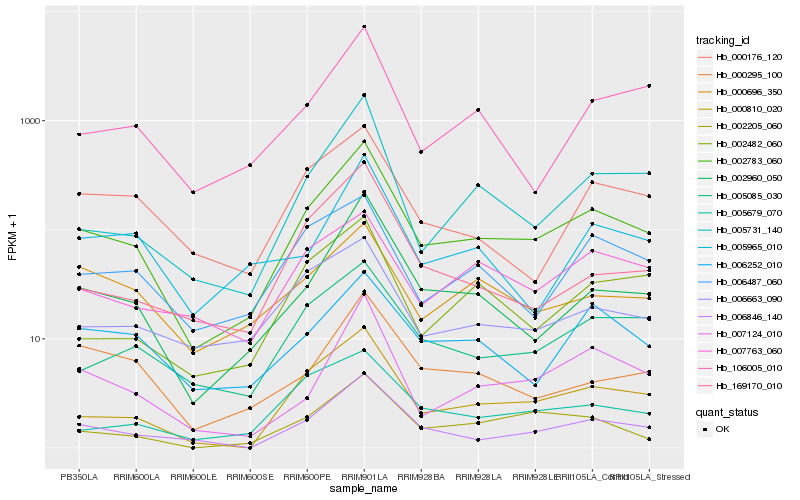
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002783_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635325 [Jatropha curcas] |
| 2 |
Hb_000810_020 |
0.1173942424 |
- |
- |
- |
| 3 |
Hb_002960_050 |
0.1283809807 |
- |
- |
PREDICTED: uncharacterized protein LOC105635325 [Jatropha curcas] |
| 4 |
Hb_006846_140 |
0.1309029141 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_005085_030 |
0.1421876942 |
- |
- |
PREDICTED: N-acetyltransferase 9-like protein isoform X1 [Jatropha curcas] |
| 6 |
Hb_005731_140 |
0.1508723553 |
- |
- |
hypothetical protein JCGZ_26808 [Jatropha curcas] |
| 7 |
Hb_007124_010 |
0.1514256603 |
- |
- |
unnamed protein product [Coffea canephora] |
| 8 |
Hb_002205_060 |
0.1563486767 |
- |
- |
- |
| 9 |
Hb_005679_070 |
0.158881964 |
- |
- |
hypothetical protein M569_07741, partial [Genlisea aurea] |
| 10 |
Hb_006663_090 |
0.1599846844 |
- |
- |
hypothetical protein B456_010G1255001, partial [Gossypium raimondii] |
| 11 |
Hb_002482_060 |
0.1611903055 |
- |
- |
hypothetical protein CICLE_v10033884mg, partial [Citrus clementina] |
| 12 |
Hb_006487_060 |
0.1618316273 |
- |
- |
Defective in cullin neddylation protein, putative [Ricinus communis] |
| 13 |
Hb_106005_010 |
0.1632413618 |
- |
- |
EG5651 [Manihot esculenta] |
| 14 |
Hb_000176_120 |
0.16603666 |
- |
- |
seryl-tRNA synthetase, putative [Ricinus communis] |
| 15 |
Hb_007763_060 |
0.1664977775 |
- |
- |
BnaA02g23510D [Brassica napus] |
| 16 |
Hb_000295_100 |
0.1671533059 |
- |
- |
- |
| 17 |
Hb_006252_010 |
0.1679291826 |
- |
- |
PREDICTED: alpha-ketoglutarate-dependent dioxygenase alkB homolog 6 isoform X1 [Jatropha curcas] |
| 18 |
Hb_005965_010 |
0.1728236361 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb family transcription factor APL-like [Jatropha curcas] |
| 19 |
Hb_169170_010 |
0.1733121281 |
- |
- |
PREDICTED: protein disulfide-isomerase 5-1 isoform X2 [Nelumbo nucifera] |
| 20 |
Hb_000696_350 |
0.1760185972 |
- |
- |
ARM repeat superfamily protein isoform 2 [Theobroma cacao] |