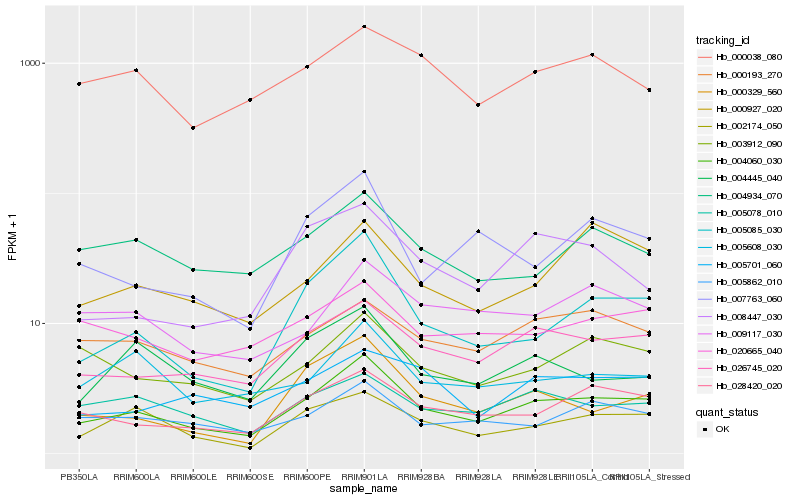
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004060_030 |
0.0 |
- |
- |
PREDICTED: primase homolog protein [Jatropha curcas] |
| 2 |
Hb_026745_020 |
0.1078915581 |
- |
- |
PREDICTED: rhodanese-like domain-containing protein 8, chloroplastic [Jatropha curcas] |
| 3 |
Hb_005085_030 |
0.1218977311 |
- |
- |
PREDICTED: N-acetyltransferase 9-like protein isoform X1 [Jatropha curcas] |
| 4 |
Hb_028420_020 |
0.1239449882 |
- |
- |
PREDICTED: uncharacterized protein LOC105647566 [Jatropha curcas] |
| 5 |
Hb_002174_050 |
0.1373003219 |
- |
- |
- |
| 6 |
Hb_005862_010 |
0.1413912875 |
- |
- |
PREDICTED: riboflavin biosynthesis protein PYRD, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000329_560 |
0.1437439853 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_003912_090 |
0.1452507327 |
- |
- |
peptidyl-tRNA hydrolase, putative [Ricinus communis] |
| 9 |
Hb_005078_010 |
0.1466351033 |
- |
- |
Derlin-2, putative [Ricinus communis] |
| 10 |
Hb_005608_030 |
0.1467524181 |
- |
- |
PREDICTED: uncharacterized protein LOC104110884 [Nicotiana tomentosiformis] |
| 11 |
Hb_000193_270 |
0.148201462 |
transcription factor |
TF Family: Pseudo ARR-B |
PREDICTED: two-component response regulator-like APRR1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_009117_030 |
0.1494406189 |
- |
- |
PREDICTED: histidine protein methyltransferase 1 homolog isoform X2 [Jatropha curcas] |
| 13 |
Hb_004934_070 |
0.150057994 |
- |
- |
PREDICTED: U2 small nuclear ribonucleoprotein A' [Jatropha curcas] |
| 14 |
Hb_000038_080 |
0.1511995576 |
- |
- |
PREDICTED: glutathione S-transferase F9-like [Jatropha curcas] |
| 15 |
Hb_004445_040 |
0.1513720787 |
- |
- |
PREDICTED: uncharacterized protein LOC105639134 isoform X5 [Jatropha curcas] |
| 16 |
Hb_020665_040 |
0.1526762939 |
transcription factor |
TF Family: Jumonji |
PREDICTED: trichohyalin-like [Malus domestica] |
| 17 |
Hb_007763_060 |
0.1533713856 |
- |
- |
BnaA02g23510D [Brassica napus] |
| 18 |
Hb_000927_020 |
0.1562152864 |
- |
- |
synaptosomal associated protein, putative [Ricinus communis] |
| 19 |
Hb_005701_060 |
0.1565222805 |
- |
- |
hypothetical protein JCGZ_04513 [Jatropha curcas] |
| 20 |
Hb_008447_030 |
0.1570916168 |
- |
- |
- |