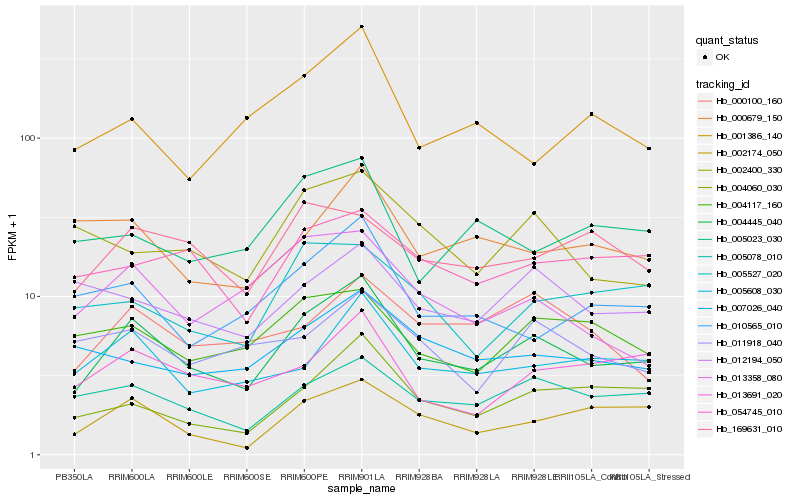
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004445_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105639134 isoform X5 [Jatropha curcas] |
| 2 |
Hb_005608_030 |
0.1423598926 |
- |
- |
PREDICTED: uncharacterized protein LOC104110884 [Nicotiana tomentosiformis] |
| 3 |
Hb_005078_010 |
0.1460880354 |
- |
- |
Derlin-2, putative [Ricinus communis] |
| 4 |
Hb_002174_050 |
0.150229849 |
- |
- |
- |
| 5 |
Hb_004060_030 |
0.1513720787 |
- |
- |
PREDICTED: primase homolog protein [Jatropha curcas] |
| 6 |
Hb_013691_020 |
0.1591442034 |
- |
- |
transcription initiation factor iie, alpha subunit, putative [Ricinus communis] |
| 7 |
Hb_010565_010 |
0.1606747725 |
- |
- |
PREDICTED: N-acetylglucosaminyl-phosphatidylinositol de-N-acetylase-like isoform X2 [Populus euphratica] |
| 8 |
Hb_013358_080 |
0.1624021084 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 25-like [Jatropha curcas] |
| 9 |
Hb_004117_160 |
0.1630926077 |
transcription factor |
TF Family: B3 |
B3 domain-containing transcription repressor VAL2 -like protein [Gossypium arboreum] |
| 10 |
Hb_000100_160 |
0.1645481649 |
- |
- |
long-chain-fatty-acid CoA ligase, putative [Ricinus communis] |
| 11 |
Hb_054745_010 |
0.165020709 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_007026_040 |
0.1680726196 |
- |
- |
PREDICTED: chaperonin 60 subunit alpha 2, chloroplastic-like [Eucalyptus grandis] |
| 13 |
Hb_169631_010 |
0.1707470008 |
- |
- |
MADS-box transcription factor, putative [Ricinus communis] |
| 14 |
Hb_005527_020 |
0.1713504555 |
- |
- |
PREDICTED: uncharacterized protein LOC105643027 [Jatropha curcas] |
| 15 |
Hb_012194_050 |
0.1720857683 |
- |
- |
Aspartyl aminopeptidase, putative [Ricinus communis] |
| 16 |
Hb_000679_150 |
0.1722932086 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 56 isoform X2 [Fragaria vesca subsp. vesca] |
| 17 |
Hb_005023_030 |
0.1732428066 |
- |
- |
PREDICTED: uncharacterized protein LOC103417254 [Malus domestica] |
| 18 |
Hb_011918_040 |
0.1734563339 |
- |
- |
PREDICTED: dnaJ protein homolog 2 [Jatropha curcas] |
| 19 |
Hb_002400_330 |
0.173506361 |
- |
- |
PREDICTED: ER membrane protein complex subunit 7 homolog isoform X2 [Jatropha curcas] |
| 20 |
Hb_001386_140 |
0.1737429636 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |