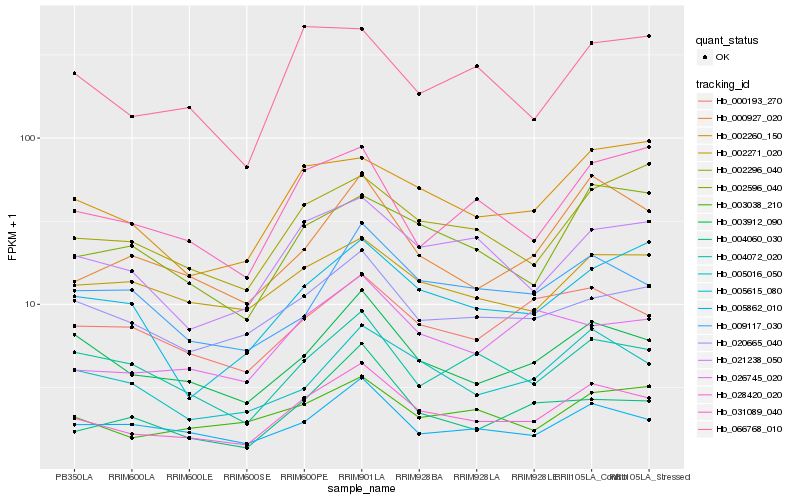
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_028420_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647566 [Jatropha curcas] |
| 2 |
Hb_003912_090 |
0.0954537205 |
- |
- |
peptidyl-tRNA hydrolase, putative [Ricinus communis] |
| 3 |
Hb_021238_050 |
0.1067673467 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_005862_010 |
0.1089007126 |
- |
- |
PREDICTED: riboflavin biosynthesis protein PYRD, chloroplastic [Jatropha curcas] |
| 5 |
Hb_020665_040 |
0.1134988599 |
transcription factor |
TF Family: Jumonji |
PREDICTED: trichohyalin-like [Malus domestica] |
| 6 |
Hb_005016_050 |
0.1142762328 |
- |
- |
- |
| 7 |
Hb_003038_210 |
0.1158214594 |
- |
- |
hypothetical protein B456_006G076400 [Gossypium raimondii] |
| 8 |
Hb_000927_020 |
0.117564449 |
- |
- |
synaptosomal associated protein, putative [Ricinus communis] |
| 9 |
Hb_002260_150 |
0.1191514058 |
- |
- |
PREDICTED: uncharacterized protein LOC105647187 isoform X2 [Jatropha curcas] |
| 10 |
Hb_002596_040 |
0.1203776337 |
- |
- |
PREDICTED: uncharacterized protein LOC105645264 isoform X2 [Jatropha curcas] |
| 11 |
Hb_002296_040 |
0.1210759672 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit 4 [Jatropha curcas] |
| 12 |
Hb_026745_020 |
0.121669423 |
- |
- |
PREDICTED: rhodanese-like domain-containing protein 8, chloroplastic [Jatropha curcas] |
| 13 |
Hb_004060_030 |
0.1239449882 |
- |
- |
PREDICTED: primase homolog protein [Jatropha curcas] |
| 14 |
Hb_000193_270 |
0.124083734 |
transcription factor |
TF Family: Pseudo ARR-B |
PREDICTED: two-component response regulator-like APRR1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_066768_010 |
0.1245257893 |
- |
- |
PREDICTED: outer envelope pore protein 16-3, chloroplastic/mitochondrial [Jatropha curcas] |
| 16 |
Hb_004072_020 |
0.1248446697 |
- |
- |
PREDICTED: uncharacterized protein LOC105635274 [Jatropha curcas] |
| 17 |
Hb_009117_030 |
0.1251899644 |
- |
- |
PREDICTED: histidine protein methyltransferase 1 homolog isoform X2 [Jatropha curcas] |
| 18 |
Hb_031089_040 |
0.1267372172 |
- |
- |
thioredoxin H-type 1 [Hevea brasiliensis] |
| 19 |
Hb_005615_080 |
0.1269050601 |
- |
- |
PREDICTED: uncharacterized protein LOC105649609 [Jatropha curcas] |
| 20 |
Hb_002271_020 |
0.1270069418 |
- |
- |
hypothetical protein EUGRSUZ_G02641 [Eucalyptus grandis] |