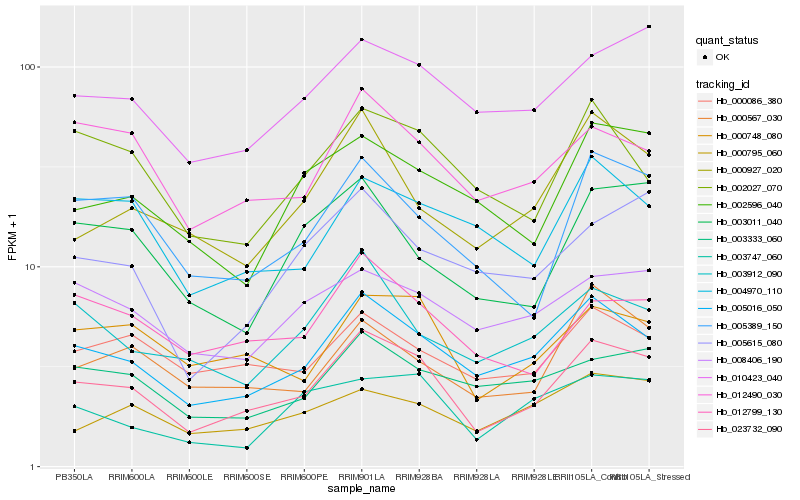
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_023732_090 |
0.0 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 2 |
Hb_005016_050 |
0.0939341855 |
- |
- |
- |
| 3 |
Hb_000748_080 |
0.1175278666 |
- |
- |
hypothetical protein OsI_28173 [Oryza sativa Indica Group] |
| 4 |
Hb_005389_150 |
0.1240975892 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_010423_040 |
0.1261507587 |
- |
- |
hypothetical protein JCGZ_17463 [Jatropha curcas] |
| 6 |
Hb_012799_130 |
0.1292372743 |
- |
- |
PREDICTED: flap endonuclease GEN-like 2 [Jatropha curcas] |
| 7 |
Hb_012490_030 |
0.1313013973 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 2 isoform X1 [Jatropha curcas] |
| 8 |
Hb_003333_060 |
0.1381893035 |
- |
- |
PREDICTED: uncharacterized protein LOC105628461 [Jatropha curcas] |
| 9 |
Hb_000567_030 |
0.1387874098 |
- |
- |
hypothetical protein JCGZ_21550 [Jatropha curcas] |
| 10 |
Hb_000086_380 |
0.1389818374 |
- |
- |
PREDICTED: succinyl-CoA ligase [ADP-forming] subunit beta, mitochondrial [Jatropha curcas] |
| 11 |
Hb_003912_090 |
0.1401845924 |
- |
- |
peptidyl-tRNA hydrolase, putative [Ricinus communis] |
| 12 |
Hb_008406_190 |
0.1408211763 |
- |
- |
PREDICTED: RNA polymerase II subunit 5-mediating protein homolog isoform X2 [Jatropha curcas] |
| 13 |
Hb_000927_020 |
0.1418560826 |
- |
- |
synaptosomal associated protein, putative [Ricinus communis] |
| 14 |
Hb_003011_040 |
0.1420371069 |
- |
- |
PREDICTED: uncharacterized protein LOC102629536 isoform X2 [Citrus sinensis] |
| 15 |
Hb_005615_080 |
0.1420585037 |
- |
- |
PREDICTED: uncharacterized protein LOC105649609 [Jatropha curcas] |
| 16 |
Hb_002027_070 |
0.1421002639 |
- |
- |
Auxin-binding protein T85 precursor, putative [Ricinus communis] |
| 17 |
Hb_003747_060 |
0.1435907991 |
- |
- |
PREDICTED: CMP-N-acetylneuraminate-beta-galactosamide-alpha-2,3-sialyltransferase 2 isoform X2 [Cucumis sativus] |
| 18 |
Hb_002596_040 |
0.1443766832 |
- |
- |
PREDICTED: uncharacterized protein LOC105645264 isoform X2 [Jatropha curcas] |
| 19 |
Hb_004970_110 |
0.1449583755 |
- |
- |
PREDICTED: protein ROOT INITIATION DEFECTIVE 3 [Jatropha curcas] |
| 20 |
Hb_000795_060 |
0.1461425202 |
rubber biosynthesis |
Gene Name: Acetyl-CoA acetyltransferase |
acetyl-CoA C-acetyltransferase [Hevea brasiliensis] |