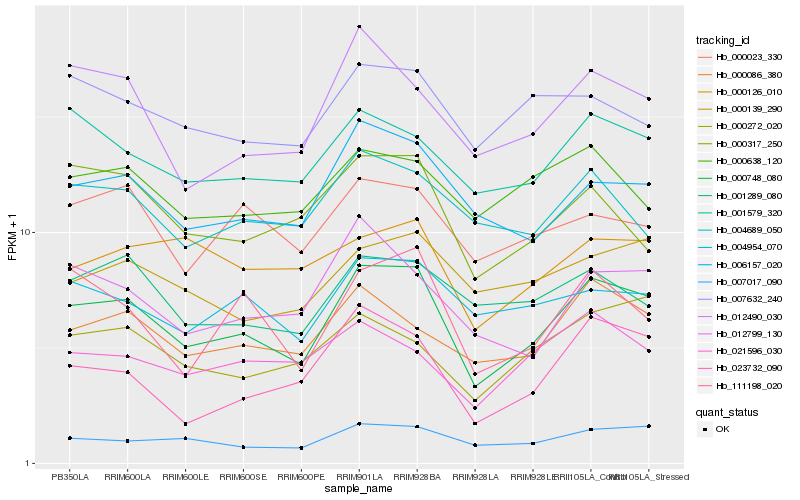
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000748_080 |
0.0 |
- |
- |
hypothetical protein OsI_28173 [Oryza sativa Indica Group] |
| 2 |
Hb_006157_020 |
0.1036264098 |
- |
- |
PREDICTED: nucleolin 1-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_007017_090 |
0.1049215113 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 186 [Jatropha curcas] |
| 4 |
Hb_000086_380 |
0.1084180902 |
- |
- |
PREDICTED: succinyl-CoA ligase [ADP-forming] subunit beta, mitochondrial [Jatropha curcas] |
| 5 |
Hb_012490_030 |
0.1118360758 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001579_320 |
0.1122619797 |
- |
- |
pyroglutamyl-peptidase I, putative [Ricinus communis] |
| 7 |
Hb_111198_020 |
0.112520778 |
- |
- |
hypothetical protein JCGZ_06907 [Jatropha curcas] |
| 8 |
Hb_012799_130 |
0.1166778992 |
- |
- |
PREDICTED: flap endonuclease GEN-like 2 [Jatropha curcas] |
| 9 |
Hb_004954_070 |
0.1173849947 |
- |
- |
PREDICTED: uncharacterized protein LOC105645187 [Jatropha curcas] |
| 10 |
Hb_023732_090 |
0.1175278666 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 11 |
Hb_000023_330 |
0.1212828184 |
- |
- |
RNA recognition motif-containing family protein [Populus trichocarpa] |
| 12 |
Hb_000139_290 |
0.1223435388 |
- |
- |
PREDICTED: peroxisomal adenine nucleotide carrier 1-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_021596_030 |
0.1231580876 |
- |
- |
PREDICTED: uncharacterized protein LOC105631841 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001289_080 |
0.1232168797 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 15 |
Hb_000272_020 |
0.1237354932 |
- |
- |
oxidoreductase, putative [Ricinus communis] |
| 16 |
Hb_000317_250 |
0.1241386618 |
- |
- |
PREDICTED: calcium-dependent protein kinase SK5 isoform X1 [Jatropha curcas] |
| 17 |
Hb_004689_050 |
0.1247441425 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000638_120 |
0.1272023926 |
- |
- |
PREDICTED: 2-oxoisovalerate dehydrogenase subunit alpha 2, mitochondrial-like [Jatropha curcas] |
| 19 |
Hb_007632_240 |
0.1272741217 |
- |
- |
transporter, putative [Ricinus communis] |
| 20 |
Hb_000126_010 |
0.1275719019 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase RFK1 isoform X1 [Jatropha curcas] |