| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
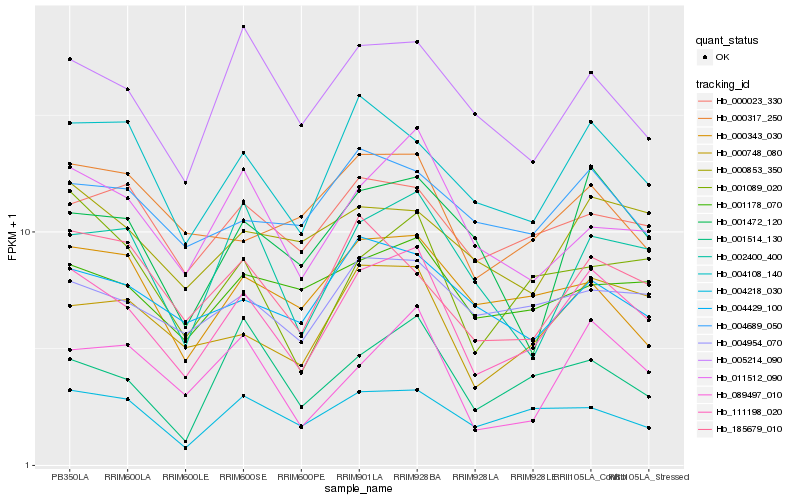
Hb_111198_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_06907 [Jatropha curcas] |
| 2 |
Hb_000748_080 |
0.112520778 |
- |
- |
hypothetical protein OsI_28173 [Oryza sativa Indica Group] |
| 3 |
Hb_004108_140 |
0.1178541902 |
- |
- |
PREDICTED: HUA2-like protein 3 isoform X3 [Jatropha curcas] |
| 4 |
Hb_011512_090 |
0.1195235373 |
- |
- |
Hydroxyethylthiazole kinase, putative [Ricinus communis] |
| 5 |
Hb_089497_010 |
0.1225857305 |
- |
- |
PREDICTED: protein EARLY RESPONSIVE TO DEHYDRATION 15-like isoform X3 [Jatropha curcas] |
| 6 |
Hb_002400_400 |
0.12572949 |
- |
- |
PREDICTED: topless-related protein 1-like isoform X3 [Jatropha curcas] |
| 7 |
Hb_001089_020 |
0.1282437415 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 2 [Jatropha curcas] |
| 8 |
Hb_004218_030 |
0.1289192709 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_005214_090 |
0.1296484284 |
- |
- |
PREDICTED: uncharacterized protein LOC105636015 [Jatropha curcas] |
| 10 |
Hb_004954_070 |
0.1317489457 |
- |
- |
PREDICTED: uncharacterized protein LOC105645187 [Jatropha curcas] |
| 11 |
Hb_001178_070 |
0.1337517177 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein FBL11 [Jatropha curcas] |
| 12 |
Hb_001472_120 |
0.1358066098 |
- |
- |
- |
| 13 |
Hb_004429_100 |
0.1358532198 |
- |
- |
PREDICTED: asparagine synthetase domain-containing protein 1 [Jatropha curcas] |
| 14 |
Hb_000023_330 |
0.1361843322 |
- |
- |
RNA recognition motif-containing family protein [Populus trichocarpa] |
| 15 |
Hb_185679_010 |
0.1362414379 |
- |
- |
serine/threonine-protein kinase rio2, putative [Ricinus communis] |
| 16 |
Hb_001514_130 |
0.138523917 |
- |
- |
hypothetical protein JCGZ_16073 [Jatropha curcas] |
| 17 |
Hb_000853_350 |
0.1405421673 |
- |
- |
PREDICTED: heparan-alpha-glucosaminide N-acetyltransferase-like [Jatropha curcas] |
| 18 |
Hb_000343_030 |
0.1407581598 |
- |
- |
PREDICTED: uncharacterized protein LOC105634976 [Jatropha curcas] |
| 19 |
Hb_004689_050 |
0.1422841412 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000317_250 |
0.1430310317 |
- |
- |
PREDICTED: calcium-dependent protein kinase SK5 isoform X1 [Jatropha curcas] |