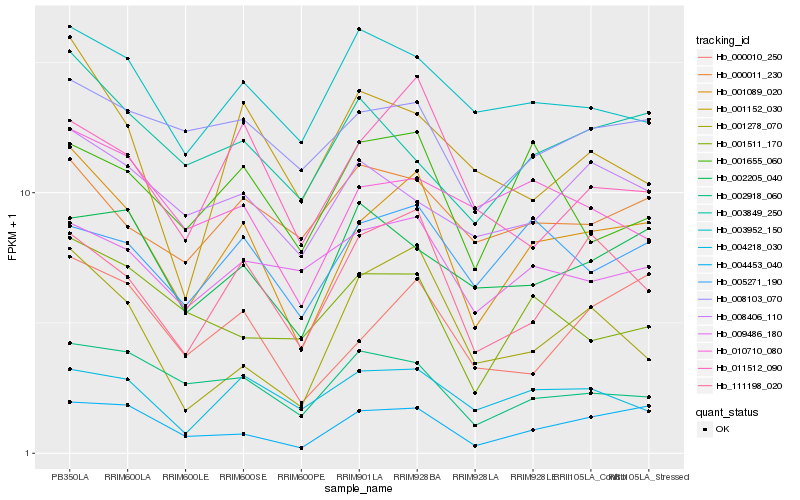
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001089_020 |
0.0 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 2 [Jatropha curcas] |
| 2 |
Hb_000010_250 |
0.1225207069 |
- |
- |
PREDICTED: protein tipD [Jatropha curcas] |
| 3 |
Hb_111198_020 |
0.1282437415 |
- |
- |
hypothetical protein JCGZ_06907 [Jatropha curcas] |
| 4 |
Hb_004453_040 |
0.130386217 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Pyrus x bretschneideri] |
| 5 |
Hb_005271_190 |
0.1447146342 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 6 |
Hb_002918_060 |
0.1452643464 |
- |
- |
PREDICTED: glyoxysomal processing protease, glyoxysomal isoform X1 [Jatropha curcas] |
| 7 |
Hb_011512_090 |
0.1480756663 |
- |
- |
Hydroxyethylthiazole kinase, putative [Ricinus communis] |
| 8 |
Hb_008103_070 |
0.1485131371 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 4-like [Jatropha curcas] |
| 9 |
Hb_010710_080 |
0.1499259048 |
- |
- |
- |
| 10 |
Hb_003849_250 |
0.1501127781 |
- |
- |
PREDICTED: protein MAK16 homolog [Jatropha curcas] |
| 11 |
Hb_004218_030 |
0.1520543909 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001278_070 |
0.1524830173 |
- |
- |
hypothetical protein glysoja_020139 [Glycine soja] |
| 13 |
Hb_009486_180 |
0.1526850813 |
- |
- |
unknown [Glycine max] |
| 14 |
Hb_003952_150 |
0.1527702515 |
- |
- |
PREDICTED: cactin [Jatropha curcas] |
| 15 |
Hb_001655_060 |
0.1551463971 |
- |
- |
PREDICTED: RNA-binding protein 28 [Jatropha curcas] |
| 16 |
Hb_008406_110 |
0.1564551954 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RING1a isoform X1 [Jatropha curcas] |
| 17 |
Hb_000011_230 |
0.1570459486 |
- |
- |
PREDICTED: bifunctional protein FolD 4, chloroplastic-like [Jatropha curcas] |
| 18 |
Hb_002205_040 |
0.1574453887 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 19 |
Hb_001152_030 |
0.1584747707 |
- |
- |
hypothetical protein POPTR_0002s22050g [Populus trichocarpa] |
| 20 |
Hb_001511_170 |
0.1589069696 |
- |
- |
PREDICTED: GDP-Man:Man(3)GlcNAc(2)-PP-Dol alpha-1,2-mannosyltransferase [Jatropha curcas] |