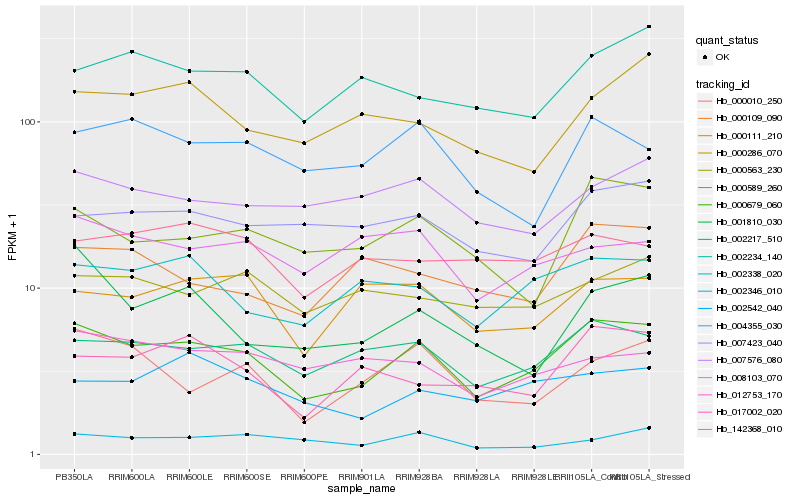
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000679_060 |
0.0 |
- |
- |
ADP,ATP carrier protein, putative [Ricinus communis] |
| 2 |
Hb_002217_510 |
0.1080033297 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002338_020 |
0.1285566034 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor SR34A [Jatropha curcas] |
| 4 |
Hb_000589_260 |
0.1321652258 |
- |
- |
3-hydroxybutyryl-CoA dehydratase, putative [Ricinus communis] |
| 5 |
Hb_007423_040 |
0.1393295807 |
- |
- |
PREDICTED: polyadenylate-binding protein 2-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_017002_020 |
0.1393419955 |
- |
- |
PREDICTED: uncharacterized protein LOC105650411 [Jatropha curcas] |
| 7 |
Hb_000010_250 |
0.1409442114 |
- |
- |
PREDICTED: protein tipD [Jatropha curcas] |
| 8 |
Hb_000111_210 |
0.141189576 |
- |
- |
PREDICTED: thioredoxin-like protein CITRX, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000109_090 |
0.1420378893 |
- |
- |
PREDICTED: deoxyhypusine hydroxylase [Jatropha curcas] |
| 10 |
Hb_002234_140 |
0.142183942 |
- |
- |
DNA-directed RNA polymerase II family protein [Populus trichocarpa] |
| 11 |
Hb_007576_080 |
0.1432318579 |
- |
- |
PREDICTED: uncharacterized protein LOC105643972 [Jatropha curcas] |
| 12 |
Hb_004355_030 |
0.1438636881 |
- |
- |
fumarylacetoacetate hydrolase, putative [Ricinus communis] |
| 13 |
Hb_002542_040 |
0.1458644221 |
- |
- |
hypothetical protein JCGZ_13884 [Jatropha curcas] |
| 14 |
Hb_000286_070 |
0.1469943165 |
- |
- |
PREDICTED: 60S ribosomal protein L13-1-like [Jatropha curcas] |
| 15 |
Hb_001810_030 |
0.1477773913 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 16 |
Hb_002346_010 |
0.1482868396 |
- |
- |
- |
| 17 |
Hb_008103_070 |
0.1493653107 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 4-like [Jatropha curcas] |
| 18 |
Hb_000563_230 |
0.1495922479 |
- |
- |
phospholipase d zeta, putative [Ricinus communis] |
| 19 |
Hb_012753_170 |
0.149990186 |
- |
- |
PREDICTED: phosphatidylinositol glycan anchor biosynthesis class U protein-like isoform X2 [Jatropha curcas] |
| 20 |
Hb_142368_010 |
0.1526450886 |
- |
- |
mRNA (guanine-7-)methyltransferase, putative [Ricinus communis] |