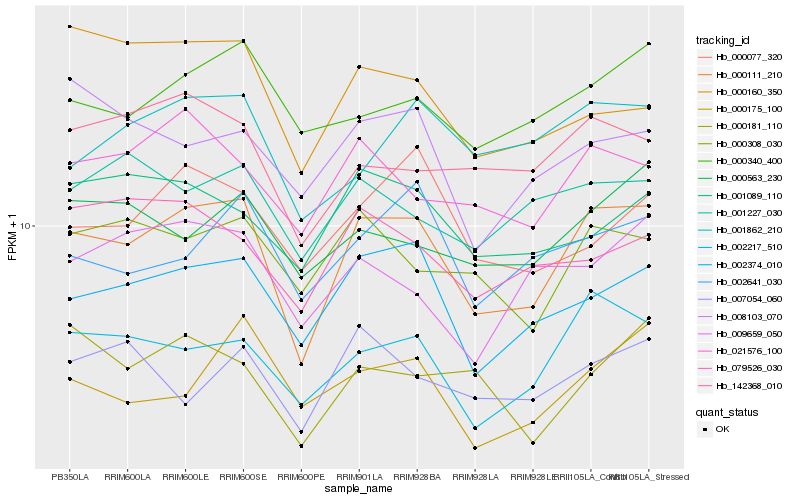
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000111_210 |
0.0 |
- |
- |
PREDICTED: thioredoxin-like protein CITRX, chloroplastic [Jatropha curcas] |
| 2 |
Hb_002374_010 |
0.0818449349 |
- |
- |
PREDICTED: protein translocase subunit SECA2, chloroplastic [Jatropha curcas] |
| 3 |
Hb_002217_510 |
0.0854471217 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_021576_100 |
0.0988069095 |
- |
- |
Enoyl-CoA hydratase, mitochondrial precursor, putative [Ricinus communis] |
| 5 |
Hb_000308_030 |
0.1016541586 |
- |
- |
PREDICTED: uncharacterized protein LOC105639686 [Jatropha curcas] |
| 6 |
Hb_000340_400 |
0.1023573469 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 7 |
Hb_142368_010 |
0.1025629339 |
- |
- |
mRNA (guanine-7-)methyltransferase, putative [Ricinus communis] |
| 8 |
Hb_001862_210 |
0.1037601891 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: salutaridine reductase-like [Citrus sinensis] |
| 9 |
Hb_002641_030 |
0.1048126752 |
- |
- |
PREDICTED: flowering time control protein FCA isoform X1 [Jatropha curcas] |
| 10 |
Hb_001089_110 |
0.1061852159 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000563_230 |
0.1064000441 |
- |
- |
phospholipase d zeta, putative [Ricinus communis] |
| 12 |
Hb_000077_320 |
0.1065331927 |
- |
- |
PREDICTED: lanC-like protein GCL2 [Jatropha curcas] |
| 13 |
Hb_001227_030 |
0.1070169628 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 2 isoform X1 [Jatropha curcas] |
| 14 |
Hb_009659_050 |
0.1077219246 |
- |
- |
PREDICTED: putative hydrolase C777.06c isoform X2 [Jatropha curcas] |
| 15 |
Hb_000175_100 |
0.108473401 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 16 |
Hb_000181_110 |
0.1091805422 |
- |
- |
PREDICTED: uncharacterized protein LOC105637521 [Jatropha curcas] |
| 17 |
Hb_079526_030 |
0.1092329208 |
- |
- |
PREDICTED: uncharacterized protein LOC105638120 [Jatropha curcas] |
| 18 |
Hb_008103_070 |
0.1104360009 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 4-like [Jatropha curcas] |
| 19 |
Hb_007054_060 |
0.1108675698 |
- |
- |
- |
| 20 |
Hb_000160_350 |
0.1116837888 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 4 isoform X3 [Vitis vinifera] |