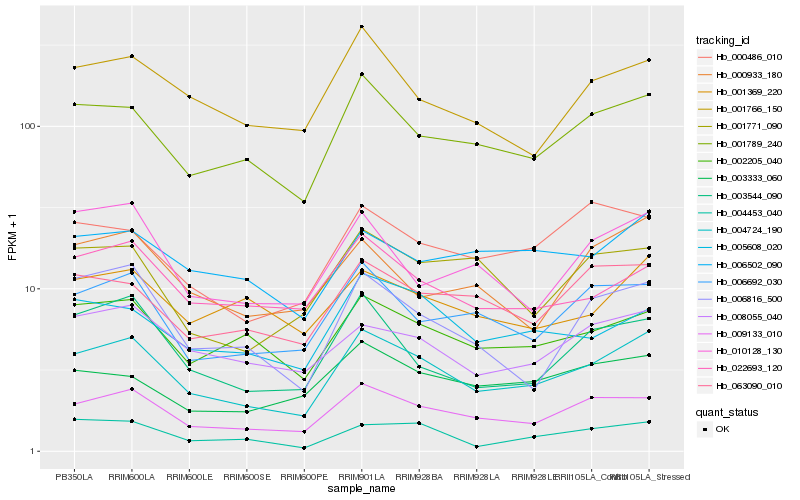
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004724_190 |
0.0 |
- |
- |
NITRILASE-LIKE protein 1 [Populus trichocarpa] |
| 2 |
Hb_009133_010 |
0.078934445 |
- |
- |
PREDICTED: L-aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase-like [Jatropha curcas] |
| 3 |
Hb_001789_240 |
0.0895842839 |
- |
- |
hypothetical protein CICLE_v10032221mg [Citrus clementina] |
| 4 |
Hb_006692_030 |
0.1097450324 |
- |
- |
PREDICTED: uncharacterized protein LOC105632913 [Jatropha curcas] |
| 5 |
Hb_002205_040 |
0.1124775146 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 6 |
Hb_008055_040 |
0.1158587647 |
- |
- |
PREDICTED: chromatin assembly factor 1 subunit FAS2 [Jatropha curcas] |
| 7 |
Hb_006816_500 |
0.1164459004 |
- |
- |
PREDICTED: splicing regulatory glutamine/lysine-rich protein 1 [Jatropha curcas] |
| 8 |
Hb_063090_010 |
0.1185149408 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 isoform X2 [Jatropha curcas] |
| 9 |
Hb_001766_150 |
0.1190260526 |
- |
- |
PREDICTED: 40S ribosomal protein S6 [Jatropha curcas] |
| 10 |
Hb_005608_020 |
0.1193533765 |
- |
- |
PREDICTED: nucleolar complex protein 2 homolog isoform X1 [Jatropha curcas] |
| 11 |
Hb_022693_120 |
0.1195795921 |
- |
- |
PREDICTED: serine/threonine-protein kinase rio2-like [Jatropha curcas] |
| 12 |
Hb_003544_090 |
0.1200811323 |
- |
- |
PREDICTED: protein HIRA isoform X2 [Nelumbo nucifera] |
| 13 |
Hb_004453_040 |
0.1258615819 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Pyrus x bretschneideri] |
| 14 |
Hb_000933_180 |
0.1265596739 |
- |
- |
PREDICTED: chorismate mutase 3, chloroplastic [Jatropha curcas] |
| 15 |
Hb_010128_130 |
0.1265789257 |
- |
- |
PREDICTED: G patch domain-containing protein 8 isoform X1 [Jatropha curcas] |
| 16 |
Hb_003333_060 |
0.1270604518 |
- |
- |
PREDICTED: uncharacterized protein LOC105628461 [Jatropha curcas] |
| 17 |
Hb_001369_220 |
0.1280707432 |
- |
- |
PREDICTED: structural maintenance of chromosomes protein 6B-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_000486_010 |
0.1282911133 |
- |
- |
PREDICTED: uncharacterized protein LOC105646661 isoform X1 [Jatropha curcas] |
| 19 |
Hb_006502_090 |
0.1285294263 |
- |
- |
PREDICTED: mRNA turnover protein 4 homolog [Jatropha curcas] |
| 20 |
Hb_001771_090 |
0.1288337102 |
- |
- |
PREDICTED: uncharacterized protein LOC105634551 [Jatropha curcas] |