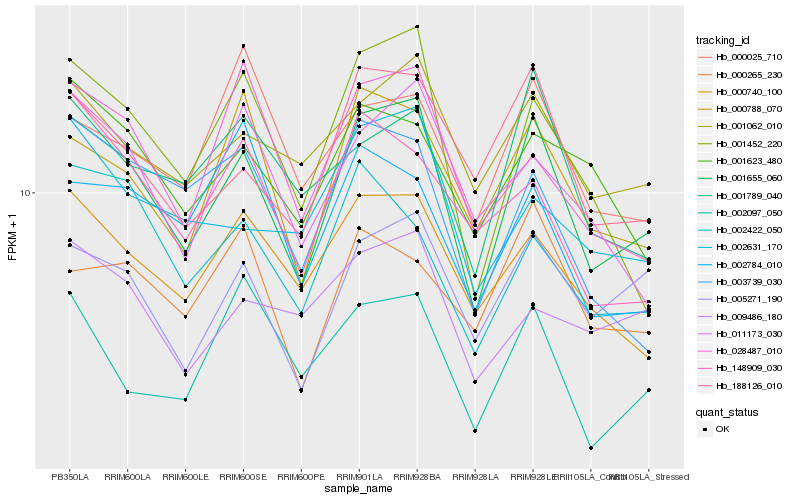
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001655_060 |
0.0 |
- |
- |
PREDICTED: RNA-binding protein 28 [Jatropha curcas] |
| 2 |
Hb_005271_190 |
0.0828241522 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 3 |
Hb_000788_070 |
0.0829667997 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g10270 [Jatropha curcas] |
| 4 |
Hb_188126_010 |
0.0842628569 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25-like isoform X3 [Jatropha curcas] |
| 5 |
Hb_000740_100 |
0.0910068017 |
- |
- |
calpain, putative [Ricinus communis] |
| 6 |
Hb_001062_010 |
0.0912841281 |
- |
- |
PREDICTED: CWF19-like protein 2 [Jatropha curcas] |
| 7 |
Hb_001789_040 |
0.091769514 |
- |
- |
PREDICTED: uncharacterized protein LOC105648457 isoform X2 [Jatropha curcas] |
| 8 |
Hb_001452_220 |
0.0933731302 |
- |
- |
PREDICTED: molybdopterin biosynthesis protein CNX1 [Jatropha curcas] |
| 9 |
Hb_002422_050 |
0.0961524863 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 5 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002097_050 |
0.0969076392 |
- |
- |
PREDICTED: protein fluG isoform X1 [Jatropha curcas] |
| 11 |
Hb_028487_010 |
0.0975253399 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 23 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000265_230 |
0.0999081264 |
transcription factor |
TF Family: HB |
PREDICTED: uncharacterized protein LOC105643603 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002631_170 |
0.1010586032 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 36 [Jatropha curcas] |
| 14 |
Hb_001623_480 |
0.101874208 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 13 [Jatropha curcas] |
| 15 |
Hb_003739_030 |
0.1027123285 |
- |
- |
PREDICTED: putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX16 [Jatropha curcas] |
| 16 |
Hb_011173_030 |
0.1030834929 |
- |
- |
pumilio, putative [Ricinus communis] |
| 17 |
Hb_000025_710 |
0.1032677469 |
transcription factor |
TF Family: SNF2 |
Chromodomain-helicase-DNA-binding 2 [Gossypium arboreum] |
| 18 |
Hb_002784_010 |
0.1051548136 |
- |
- |
DNA-directed RNA polymerase I subunit, putative [Ricinus communis] |
| 19 |
Hb_009486_180 |
0.1059812713 |
- |
- |
unknown [Glycine max] |
| 20 |
Hb_148909_030 |
0.1062945151 |
- |
- |
PREDICTED: uncharacterized protein LOC105638192 isoform X1 [Jatropha curcas] |