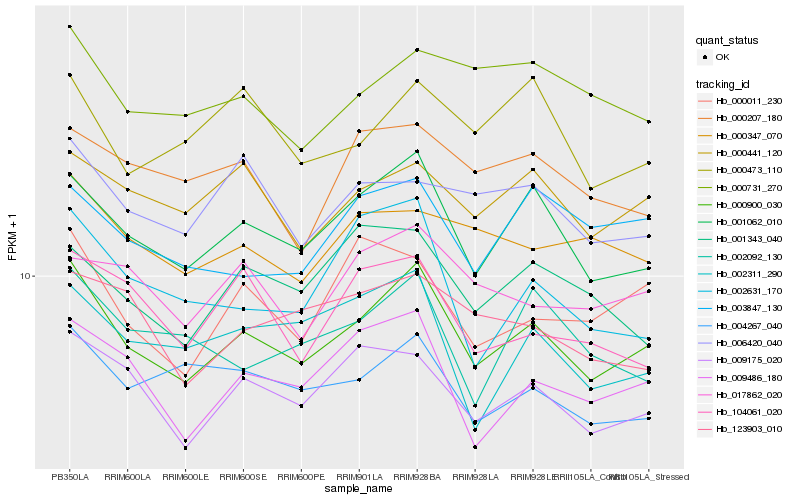
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000900_030 |
0.0 |
- |
- |
PREDICTED: WD repeat-containing protein 3 [Jatropha curcas] |
| 2 |
Hb_001062_010 |
0.0494022671 |
- |
- |
PREDICTED: CWF19-like protein 2 [Jatropha curcas] |
| 3 |
Hb_009175_020 |
0.0646454162 |
- |
- |
PREDICTED: uncharacterized protein LOC101303140 [Fragaria vesca subsp. vesca] |
| 4 |
Hb_009486_180 |
0.0665837118 |
- |
- |
unknown [Glycine max] |
| 5 |
Hb_000441_120 |
0.0748202168 |
- |
- |
PREDICTED: DNA-damage-repair/toleration protein DRT111, chloroplastic isoform X1 [Jatropha curcas] |
| 6 |
Hb_004267_040 |
0.0763490193 |
- |
- |
PREDICTED: transcriptional adapter ADA2a [Jatropha curcas] |
| 7 |
Hb_000011_230 |
0.0782829834 |
- |
- |
PREDICTED: bifunctional protein FolD 4, chloroplastic-like [Jatropha curcas] |
| 8 |
Hb_000347_070 |
0.0810740033 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002311_290 |
0.0815065144 |
- |
- |
PREDICTED: pumilio homolog 24 [Jatropha curcas] |
| 10 |
Hb_017862_020 |
0.0818981507 |
- |
- |
PREDICTED: probable inactive serine/threonine-protein kinase lvsG [Jatropha curcas] |
| 11 |
Hb_000731_270 |
0.0823329618 |
- |
- |
PREDICTED: polyadenylate-binding protein RBP45B isoform X1 [Jatropha curcas] |
| 12 |
Hb_006420_040 |
0.0824049315 |
transcription factor |
TF Family: Orphans |
PREDICTED: paired amphipathic helix protein Sin3-like 4 isoform X2 [Jatropha curcas] |
| 13 |
Hb_003847_130 |
0.0832213624 |
- |
- |
cytochrome C oxidase assembly protein cox11, putative [Ricinus communis] |
| 14 |
Hb_002631_170 |
0.0833842883 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 36 [Jatropha curcas] |
| 15 |
Hb_002092_130 |
0.0837316425 |
- |
- |
PREDICTED: protein CASP-like [Oryza brachyantha] |
| 16 |
Hb_000473_110 |
0.0840628808 |
- |
- |
PREDICTED: PP2A regulatory subunit TAP46 [Jatropha curcas] |
| 17 |
Hb_104061_020 |
0.0842347252 |
- |
- |
PREDICTED: UV-stimulated scaffold protein A homolog [Jatropha curcas] |
| 18 |
Hb_001343_040 |
0.0848413303 |
- |
- |
PREDICTED: uncharacterized protein LOC105638555 [Jatropha curcas] |
| 19 |
Hb_000207_180 |
0.08493062 |
- |
- |
PREDICTED: calponin homology domain-containing protein DDB_G0272472 [Jatropha curcas] |
| 20 |
Hb_123903_010 |
0.0849931962 |
transcription factor |
TF Family: IWS1 |
conserved hypothetical protein [Ricinus communis] |