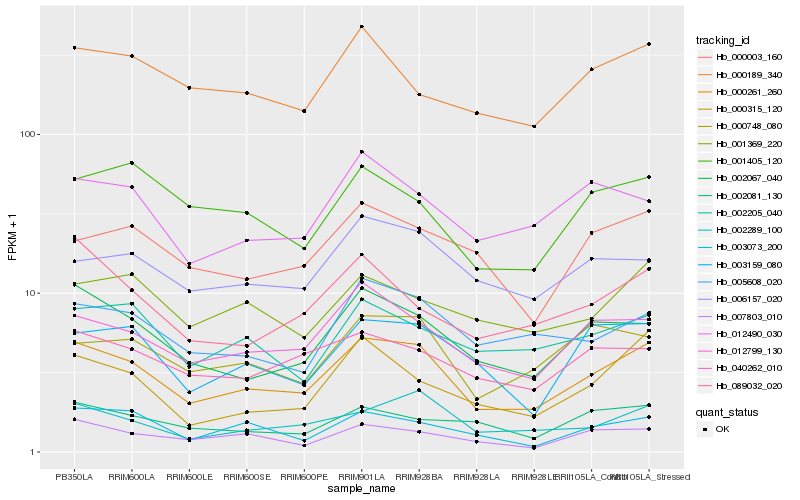
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000261_260 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: HBS1-like protein [Populus euphratica] |
| 2 |
Hb_012799_130 |
0.1081449333 |
- |
- |
PREDICTED: flap endonuclease GEN-like 2 [Jatropha curcas] |
| 3 |
Hb_002067_040 |
0.108328157 |
- |
- |
dihydrodipicolinate reductase, putative [Ricinus communis] |
| 4 |
Hb_002289_100 |
0.1157586901 |
- |
- |
PREDICTED: cytoplasmic tRNA 2-thiolation protein 2 [Jatropha curcas] |
| 5 |
Hb_003159_080 |
0.1219547405 |
- |
- |
PREDICTED: uncharacterized protein LOC105639276 [Jatropha curcas] |
| 6 |
Hb_005608_020 |
0.1250578337 |
- |
- |
PREDICTED: nucleolar complex protein 2 homolog isoform X1 [Jatropha curcas] |
| 7 |
Hb_000748_080 |
0.1277882055 |
- |
- |
hypothetical protein OsI_28173 [Oryza sativa Indica Group] |
| 8 |
Hb_007803_010 |
0.1279703495 |
- |
- |
PREDICTED: F-box protein At3g07870-like [Jatropha curcas] |
| 9 |
Hb_040262_010 |
0.1294178602 |
- |
- |
PREDICTED: aspartate carbamoyltransferase 1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_003073_200 |
0.131274329 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g11410 [Jatropha curcas] |
| 11 |
Hb_000003_160 |
0.1316490657 |
- |
- |
PREDICTED: mitochondrial import receptor subunit TOM40-1 [Jatropha curcas] |
| 12 |
Hb_000315_120 |
0.1330443934 |
- |
- |
- |
| 13 |
Hb_012490_030 |
0.1331877758 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 2 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002205_040 |
0.1331981444 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 15 |
Hb_001369_220 |
0.1336887351 |
- |
- |
PREDICTED: structural maintenance of chromosomes protein 6B-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_000189_340 |
0.133871295 |
- |
- |
PREDICTED: 40S ribosomal protein S3a-1 [Jatropha curcas] |
| 17 |
Hb_006157_020 |
0.1348092068 |
- |
- |
PREDICTED: nucleolin 1-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_002081_130 |
0.1351531197 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 19 |
Hb_001405_120 |
0.135398967 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit H [Jatropha curcas] |
| 20 |
Hb_089032_020 |
0.1355740533 |
- |
- |
PREDICTED: BRCA1-associated protein [Jatropha curcas] |