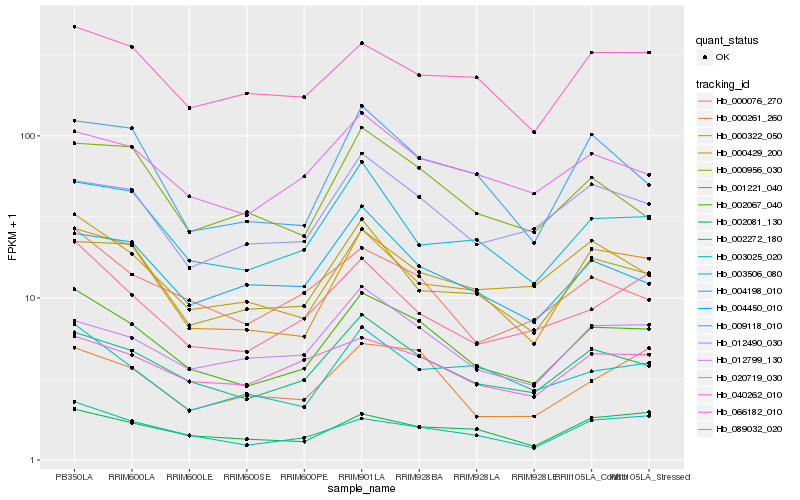
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002067_040 |
0.0 |
- |
- |
dihydrodipicolinate reductase, putative [Ricinus communis] |
| 2 |
Hb_002272_180 |
0.070637179 |
- |
- |
hypothetical protein JCGZ_23380 [Jatropha curcas] |
| 3 |
Hb_000322_050 |
0.0920100778 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 4-like [Jatropha curcas] |
| 4 |
Hb_003025_020 |
0.0932615658 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 5 |
Hb_000076_270 |
0.0976642587 |
- |
- |
PREDICTED: DDRGK domain-containing protein 1 [Populus euphratica] |
| 6 |
Hb_000429_200 |
0.1003988978 |
- |
- |
PREDICTED: DNA-binding protein SMUBP-2 [Jatropha curcas] |
| 7 |
Hb_012490_030 |
0.1008358113 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 2 isoform X1 [Jatropha curcas] |
| 8 |
Hb_020719_030 |
0.1013804696 |
- |
- |
PREDICTED: ER membrane protein complex subunit 3-like [Jatropha curcas] |
| 9 |
Hb_004198_010 |
0.1022761411 |
- |
- |
Eukaryotic translation initiation factor 2 subunit beta [Theobroma cacao] |
| 10 |
Hb_089032_020 |
0.1035588691 |
- |
- |
PREDICTED: BRCA1-associated protein [Jatropha curcas] |
| 11 |
Hb_000956_030 |
0.1044740648 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 19-like [Jatropha curcas] |
| 12 |
Hb_012799_130 |
0.106717821 |
- |
- |
PREDICTED: flap endonuclease GEN-like 2 [Jatropha curcas] |
| 13 |
Hb_000261_260 |
0.108328157 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: HBS1-like protein [Populus euphratica] |
| 14 |
Hb_003506_080 |
0.1083855813 |
- |
- |
dipeptidyl peptidase IV, putative [Ricinus communis] |
| 15 |
Hb_009118_010 |
0.1097952254 |
- |
- |
methionine aminopeptidase, putative [Ricinus communis] |
| 16 |
Hb_002081_130 |
0.1098640237 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 17 |
Hb_001221_040 |
0.1099880998 |
- |
- |
PREDICTED: nucleolar protein 56-like [Jatropha curcas] |
| 18 |
Hb_066182_010 |
0.1103542408 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit beta-like protein [Jatropha curcas] |
| 19 |
Hb_004450_010 |
0.1104894709 |
- |
- |
PREDICTED: methyltransferase-like protein 10 [Jatropha curcas] |
| 20 |
Hb_040262_010 |
0.1120353532 |
- |
- |
PREDICTED: aspartate carbamoyltransferase 1, chloroplastic [Jatropha curcas] |