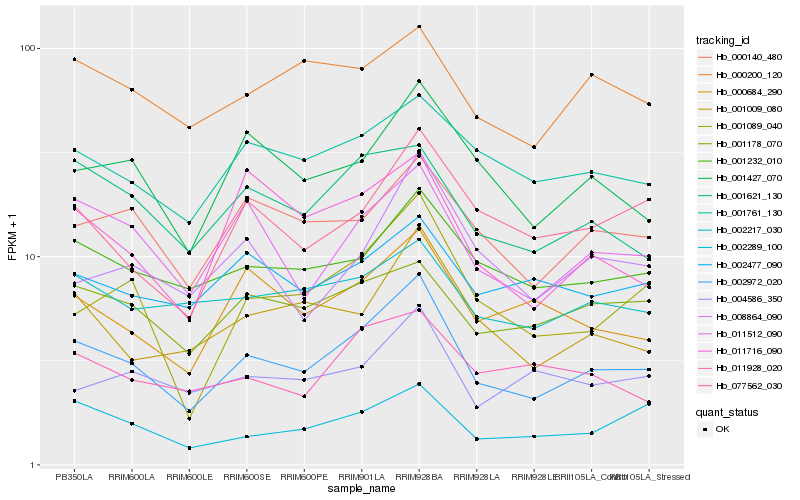
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002972_020 |
0.0 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2A isoform X1 [Jatropha curcas] |
| 2 |
Hb_011512_090 |
0.1113328648 |
- |
- |
Hydroxyethylthiazole kinase, putative [Ricinus communis] |
| 3 |
Hb_001427_070 |
0.1184480427 |
- |
- |
PREDICTED: xaa-Pro dipeptidase [Jatropha curcas] |
| 4 |
Hb_001089_040 |
0.119500045 |
transcription factor |
TF Family: bHLH |
DNA-directed RNA polymerase beta chain, putative [Ricinus communis] |
| 5 |
Hb_001009_080 |
0.1199823589 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001232_010 |
0.1201893534 |
- |
- |
PREDICTED: uncharacterized protein LOC105634976 [Jatropha curcas] |
| 7 |
Hb_000140_480 |
0.1240657108 |
- |
- |
PREDICTED: uncharacterized protein LOC105637350 [Jatropha curcas] |
| 8 |
Hb_001621_130 |
0.1251100365 |
- |
- |
PREDICTED: pumilio homolog 1-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_077562_030 |
0.1269515483 |
- |
- |
PREDICTED: cytochrome P450 4X1-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_000684_290 |
0.1274943273 |
- |
- |
ubiquitin protein ligase E3a, putative [Ricinus communis] |
| 11 |
Hb_001761_130 |
0.1280412165 |
- |
- |
PREDICTED: uncharacterized protein LOC105646118 isoform X1 [Jatropha curcas] |
| 12 |
Hb_002289_100 |
0.1346477321 |
- |
- |
PREDICTED: cytoplasmic tRNA 2-thiolation protein 2 [Jatropha curcas] |
| 13 |
Hb_011716_090 |
0.1352947725 |
transcription factor |
TF Family: HRT |
putative transciption factor family protein [Populus trichocarpa] |
| 14 |
Hb_008864_090 |
0.135371112 |
- |
- |
PREDICTED: plastid division protein PDV2-like [Jatropha curcas] |
| 15 |
Hb_001178_070 |
0.1362259466 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein FBL11 [Jatropha curcas] |
| 16 |
Hb_000200_120 |
0.1380551681 |
- |
- |
PREDICTED: probable fructose-bisphosphate aldolase 3, chloroplastic [Jatropha curcas] |
| 17 |
Hb_002217_030 |
0.1392003956 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 isoform X1 [Jatropha curcas] |
| 18 |
Hb_004586_350 |
0.1399043842 |
- |
- |
Phosphoribosylaminoimidazole-succinocarboxamide synthase [Morus notabilis] |
| 19 |
Hb_002477_090 |
0.1403265743 |
- |
- |
PREDICTED: uncharacterized protein LOC105631409 isoform X1 [Jatropha curcas] |
| 20 |
Hb_011928_020 |
0.1404270431 |
- |
- |
something about silencing protein sas10, putative [Ricinus communis] |