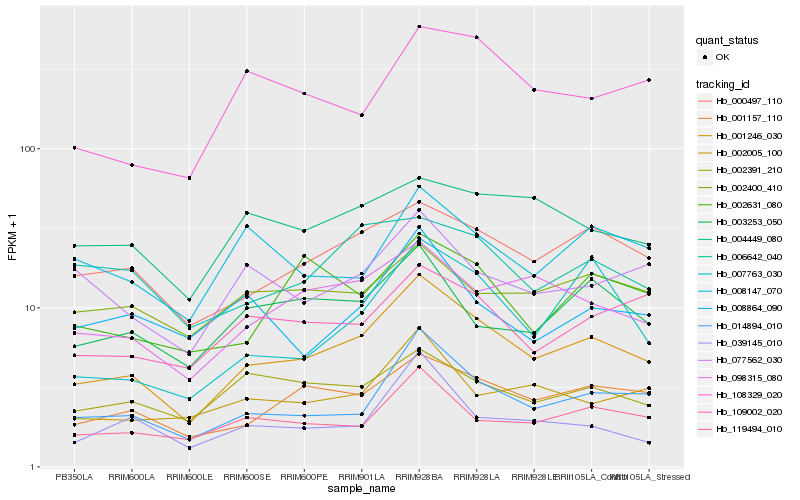
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001246_030 |
0.0 |
- |
- |
hypothetical protein JCGZ_14672 [Jatropha curcas] |
| 2 |
Hb_014894_010 |
0.100147261 |
- |
- |
hypothetical protein CICLE_v10001869mg [Citrus clementina] |
| 3 |
Hb_001157_110 |
0.1157929125 |
- |
- |
PREDICTED: formate--tetrahydrofolate ligase [Populus euphratica] |
| 4 |
Hb_000497_110 |
0.1169642521 |
- |
- |
PREDICTED: uncharacterized protein LOC105647268 [Jatropha curcas] |
| 5 |
Hb_098315_080 |
0.1253370507 |
- |
- |
methylmalonate-semialdehyde dehydrogenase, putative [Ricinus communis] |
| 6 |
Hb_119494_010 |
0.1270029235 |
- |
- |
PREDICTED: box C/D snoRNA protein 1 [Jatropha curcas] |
| 7 |
Hb_002005_100 |
0.1403855581 |
- |
- |
- |
| 8 |
Hb_003253_050 |
0.1409655159 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 3 [Jatropha curcas] |
| 9 |
Hb_109002_020 |
0.1433472766 |
- |
- |
PREDICTED: CTP synthase-like [Jatropha curcas] |
| 10 |
Hb_002391_210 |
0.1507874537 |
- |
- |
PREDICTED: putative F-box protein At1g65770 [Jatropha curcas] |
| 11 |
Hb_039145_010 |
0.1509259113 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 12 |
Hb_077562_030 |
0.1509899831 |
- |
- |
PREDICTED: cytochrome P450 4X1-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_108329_020 |
0.1518074811 |
transcription factor |
TF Family: EIL |
ETHYLENE-INSENSITIVE3 protein, putative [Ricinus communis] |
| 14 |
Hb_004449_080 |
0.1527224366 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH3-like [Jatropha curcas] |
| 15 |
Hb_006642_040 |
0.1527922421 |
- |
- |
PREDICTED: uncharacterized protein LOC105629489 [Jatropha curcas] |
| 16 |
Hb_007763_030 |
0.1535277319 |
- |
- |
PREDICTED: uncharacterized protein LOC105638154 [Jatropha curcas] |
| 17 |
Hb_002631_080 |
0.1535786687 |
- |
- |
glyceraldehyde 3-phosphate dehydrogenase, putative [Ricinus communis] |
| 18 |
Hb_008864_090 |
0.1543539671 |
- |
- |
PREDICTED: plastid division protein PDV2-like [Jatropha curcas] |
| 19 |
Hb_008147_070 |
0.1544458071 |
- |
- |
PREDICTED: tryptophan synthase beta chain 1 [Jatropha curcas] |
| 20 |
Hb_002400_410 |
0.1545285885 |
- |
- |
PREDICTED: topless-related protein 1-like [Jatropha curcas] |