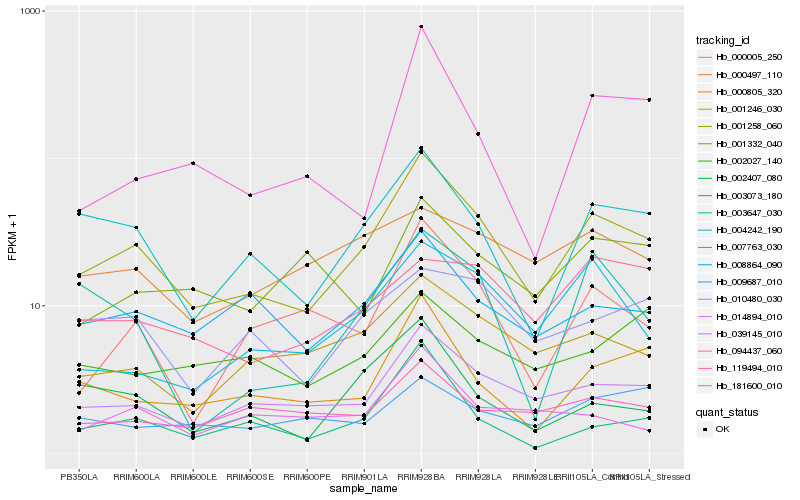
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001258_060 |
0.0 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxykinase [ATP] [Jatropha curcas] |
| 2 |
Hb_014894_010 |
0.1254735988 |
- |
- |
hypothetical protein CICLE_v10001869mg [Citrus clementina] |
| 3 |
Hb_004242_190 |
0.1332862542 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 4 |
Hb_008864_090 |
0.160941261 |
- |
- |
PREDICTED: plastid division protein PDV2-like [Jatropha curcas] |
| 5 |
Hb_001246_030 |
0.1610212923 |
- |
- |
hypothetical protein JCGZ_14672 [Jatropha curcas] |
| 6 |
Hb_000805_320 |
0.1648023413 |
- |
- |
PREDICTED: putative hydrolase C777.06c [Jatropha curcas] |
| 7 |
Hb_007763_030 |
0.1765023658 |
- |
- |
PREDICTED: uncharacterized protein LOC105638154 [Jatropha curcas] |
| 8 |
Hb_000005_250 |
0.1769928228 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_119494_010 |
0.1831981474 |
- |
- |
PREDICTED: box C/D snoRNA protein 1 [Jatropha curcas] |
| 10 |
Hb_010480_030 |
0.1835066662 |
- |
- |
PREDICTED: LETM1 and EF-hand domain-containing protein 1, mitochondrial-like [Jatropha curcas] |
| 11 |
Hb_003073_180 |
0.1863571449 |
- |
- |
hypothetical protein JCGZ_20198 [Jatropha curcas] |
| 12 |
Hb_002407_080 |
0.1863998521 |
- |
- |
hypothetical protein JCGZ_00752 [Jatropha curcas] |
| 13 |
Hb_039145_010 |
0.1930047693 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 14 |
Hb_003647_030 |
0.1947572809 |
- |
- |
PREDICTED: probable glutathione S-transferase [Populus euphratica] |
| 15 |
Hb_181600_010 |
0.1951744681 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 27 [Jatropha curcas] |
| 16 |
Hb_000497_110 |
0.1956953932 |
- |
- |
PREDICTED: uncharacterized protein LOC105647268 [Jatropha curcas] |
| 17 |
Hb_094437_060 |
0.1963313298 |
- |
- |
PREDICTED: cytochrome B5 isoform D-like [Jatropha curcas] |
| 18 |
Hb_002027_140 |
0.2016495824 |
- |
- |
PREDICTED: probable indole-3-acetic acid-amido synthetase GH3.6 [Jatropha curcas] |
| 19 |
Hb_001332_040 |
0.2017458588 |
- |
- |
PREDICTED: transcription initiation factor IIB [Fragaria vesca subsp. vesca] |
| 20 |
Hb_009687_010 |
0.2040618693 |
- |
- |
PREDICTED: protein SUPPRESSOR OF K(+) TRANSPORT GROWTH DEFECT 1-like isoform X4 [Jatropha curcas] |