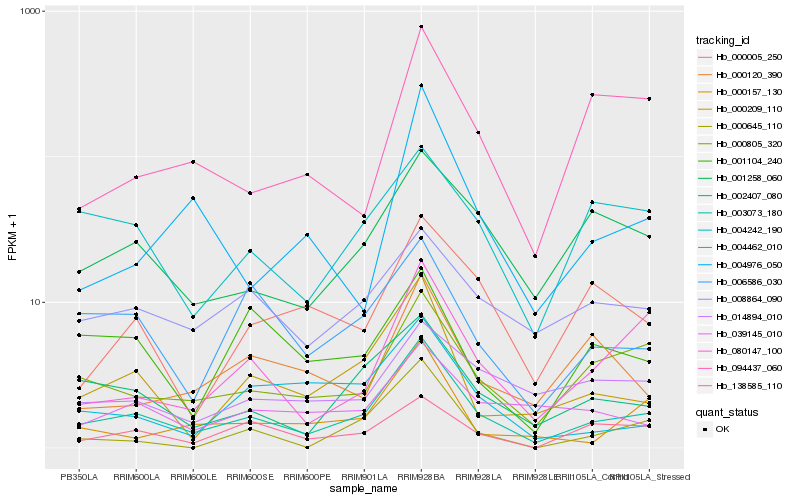
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003073_180 |
0.0 |
- |
- |
hypothetical protein JCGZ_20198 [Jatropha curcas] |
| 2 |
Hb_002407_080 |
0.167440373 |
- |
- |
hypothetical protein JCGZ_00752 [Jatropha curcas] |
| 3 |
Hb_000805_320 |
0.168723814 |
- |
- |
PREDICTED: putative hydrolase C777.06c [Jatropha curcas] |
| 4 |
Hb_000209_110 |
0.1711450568 |
- |
- |
PREDICTED: receptor-like protein kinase BRI1-like 3 [Jatropha curcas] |
| 5 |
Hb_001258_060 |
0.1863571449 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxykinase [ATP] [Jatropha curcas] |
| 6 |
Hb_080147_100 |
0.18730279 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase LIN [Jatropha curcas] |
| 7 |
Hb_006586_030 |
0.1953184498 |
- |
- |
PREDICTED: transcription factor LHW isoform X1 [Jatropha curcas] |
| 8 |
Hb_008864_090 |
0.1995850598 |
- |
- |
PREDICTED: plastid division protein PDV2-like [Jatropha curcas] |
| 9 |
Hb_004242_190 |
0.2003537414 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 10 |
Hb_138585_110 |
0.2004991246 |
- |
- |
- |
| 11 |
Hb_039145_010 |
0.2095407337 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 12 |
Hb_014894_010 |
0.2160972257 |
- |
- |
hypothetical protein CICLE_v10001869mg [Citrus clementina] |
| 13 |
Hb_000157_130 |
0.218496723 |
- |
- |
PREDICTED: cinnamoyl-CoA reductase 1-like isoform X2 [Populus euphratica] |
| 14 |
Hb_000005_250 |
0.2214113462 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_004462_010 |
0.2221763667 |
- |
- |
PREDICTED: primary amine oxidase-like [Jatropha curcas] |
| 16 |
Hb_000645_110 |
0.2223938172 |
transcription factor |
TF Family: GRAS |
PREDICTED: DELLA protein RGL1-like [Jatropha curcas] |
| 17 |
Hb_094437_060 |
0.2246743452 |
- |
- |
PREDICTED: cytochrome B5 isoform D-like [Jatropha curcas] |
| 18 |
Hb_001104_240 |
0.2255735199 |
transcription factor |
TF Family: Trihelix |
transcription factor, putative [Ricinus communis] |
| 19 |
Hb_004976_050 |
0.2315925653 |
- |
- |
- |
| 20 |
Hb_000120_390 |
0.2317955452 |
- |
- |
PREDICTED: uncharacterized transporter YBR287W-like [Jatropha curcas] |