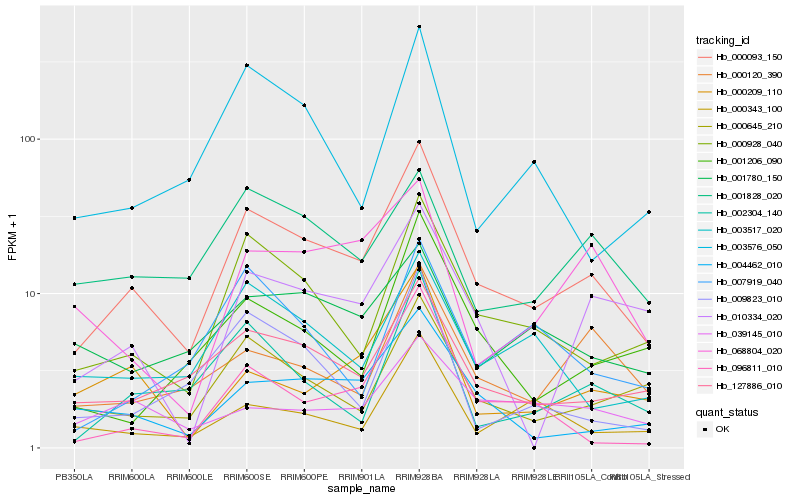
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000093_150 |
0.0 |
- |
- |
6-phosphogluconolactonase, putative [Ricinus communis] |
| 2 |
Hb_000928_040 |
0.1507526108 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 3 |
Hb_127886_010 |
0.155774441 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 4 |
Hb_000645_210 |
0.1707872639 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 5-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_004462_010 |
0.1761362433 |
- |
- |
PREDICTED: primary amine oxidase-like [Jatropha curcas] |
| 6 |
Hb_003517_020 |
0.1899784336 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter C family member 3-like [Jatropha curcas] |
| 7 |
Hb_002304_140 |
0.1931346078 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 8 |
Hb_000120_390 |
0.1938595728 |
- |
- |
PREDICTED: uncharacterized transporter YBR287W-like [Jatropha curcas] |
| 9 |
Hb_003576_050 |
0.2002819229 |
transcription factor |
TF Family: EIL |
ETHYLENE-INSENSITIVE3 protein, putative [Ricinus communis] |
| 10 |
Hb_001206_090 |
0.2003056009 |
- |
- |
pyruvate decarboxylase [Hevea brasiliensis] |
| 11 |
Hb_001828_020 |
0.2010064757 |
- |
- |
hypothetical protein CISIN_1g010132mg [Citrus sinensis] |
| 12 |
Hb_010334_020 |
0.201229705 |
- |
- |
PREDICTED: inactive protein RESTRICTED TEV MOVEMENT 1-like [Sesamum indicum] |
| 13 |
Hb_000209_110 |
0.2029997067 |
- |
- |
PREDICTED: receptor-like protein kinase BRI1-like 3 [Jatropha curcas] |
| 14 |
Hb_001780_150 |
0.2039847219 |
- |
- |
ceramidase, putative [Ricinus communis] |
| 15 |
Hb_009823_010 |
0.2066048239 |
- |
- |
PREDICTED: uncharacterized protein LOC105633096 [Jatropha curcas] |
| 16 |
Hb_039145_010 |
0.2070312003 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 17 |
Hb_096811_010 |
0.207973398 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At1g61190 [Vitis vinifera] |
| 18 |
Hb_068804_020 |
0.2120216654 |
- |
- |
hypothetical protein AMTR_s00046p00231970 [Amborella trichopoda] |
| 19 |
Hb_000343_100 |
0.2124068771 |
- |
- |
PREDICTED: regulator of telomere elongation helicase 1 homolog [Jatropha curcas] |
| 20 |
Hb_007919_040 |
0.2124237175 |
- |
- |
PREDICTED: protein LYK5 [Jatropha curcas] |