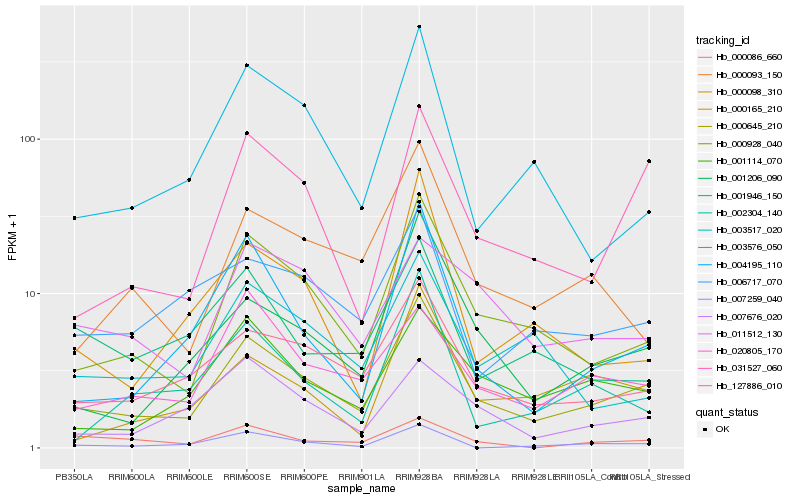
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000645_210 |
0.0 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 5-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_000928_040 |
0.1234200591 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 3 |
Hb_127886_010 |
0.1278523822 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 4 |
Hb_001206_090 |
0.1563468272 |
- |
- |
pyruvate decarboxylase [Hevea brasiliensis] |
| 5 |
Hb_031527_060 |
0.1604017123 |
- |
- |
glutamate decarboxylase, putative [Ricinus communis] |
| 6 |
Hb_001114_070 |
0.1668021208 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At1g58400 [Jatropha curcas] |
| 7 |
Hb_000093_150 |
0.1707872639 |
- |
- |
6-phosphogluconolactonase, putative [Ricinus communis] |
| 8 |
Hb_004195_110 |
0.1765320071 |
- |
- |
hypothetical protein JCGZ_24010 [Jatropha curcas] |
| 9 |
Hb_003576_050 |
0.1774316621 |
transcription factor |
TF Family: EIL |
ETHYLENE-INSENSITIVE3 protein, putative [Ricinus communis] |
| 10 |
Hb_006717_070 |
0.1788892933 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor CPC-like [Jatropha curcas] |
| 11 |
Hb_020805_170 |
0.1821377988 |
- |
- |
PREDICTED: F-box protein CPR30-like [Jatropha curcas] |
| 12 |
Hb_007259_040 |
0.1823533171 |
- |
- |
hypothetical protein VITISV_005492 [Vitis vinifera] |
| 13 |
Hb_000165_210 |
0.1854229655 |
- |
- |
PREDICTED: trihelix transcription factor ASIL2 isoform X2 [Jatropha curcas] |
| 14 |
Hb_003517_020 |
0.18644294 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter C family member 3-like [Jatropha curcas] |
| 15 |
Hb_007676_020 |
0.1875046363 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase LIN-1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001946_150 |
0.1911817612 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000098_310 |
0.1945204597 |
- |
- |
RecName: Full=Anthocyanidin 3-O-glucosyltransferase 1; AltName: Full=Flavonol 3-O-glucosyltransferase 1; AltName: Full=UDP-glucose flavonoid 3-O-glucosyltransferase 1 [Manihot esculenta] |
| 18 |
Hb_002304_140 |
0.1973086793 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 19 |
Hb_011512_130 |
0.1979047984 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At2g33170 [Jatropha curcas] |
| 20 |
Hb_000086_660 |
0.199531442 |
- |
- |
PREDICTED: phytosulfokine receptor 1-like [Jatropha curcas] |