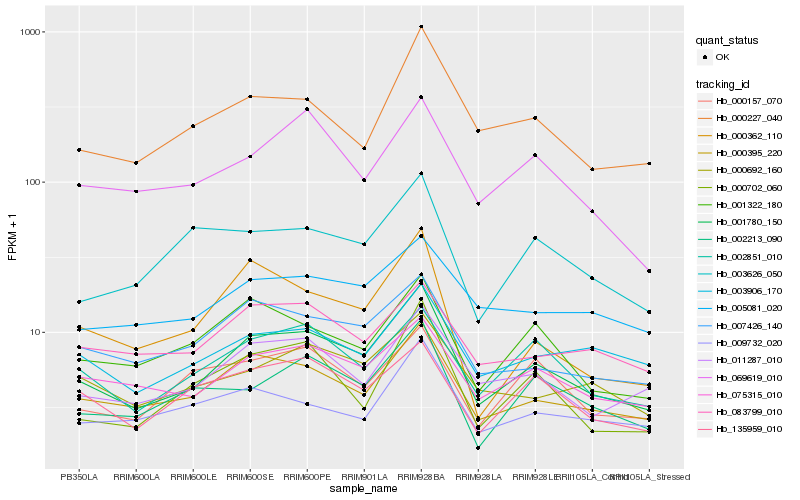
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001780_150 |
0.0 |
- |
- |
ceramidase, putative [Ricinus communis] |
| 2 |
Hb_000395_220 |
0.0885657633 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 3 |
Hb_011287_010 |
0.1091176865 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 4 |
Hb_075315_010 |
0.1114881492 |
- |
- |
ornithine aminotransferase [Camellia sinensis] |
| 5 |
Hb_007426_140 |
0.1115719919 |
- |
- |
ornithine aminotransferase, putative [Ricinus communis] |
| 6 |
Hb_002851_010 |
0.1139031558 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 7 |
Hb_000227_040 |
0.1142253926 |
- |
- |
Stem-specific protein TSJT1, putative [Ricinus communis] |
| 8 |
Hb_002213_090 |
0.1142676024 |
- |
- |
PREDICTED: uncharacterized protein LOC105640147 [Jatropha curcas] |
| 9 |
Hb_001322_180 |
0.1181114005 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 26 [Jatropha curcas] |
| 10 |
Hb_000702_060 |
0.1188615572 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000692_160 |
0.1233501131 |
- |
- |
Transportin 1 isoform 1 [Theobroma cacao] |
| 12 |
Hb_135959_010 |
0.1240071363 |
- |
- |
hypothetical protein JCGZ_07060 [Jatropha curcas] |
| 13 |
Hb_000362_110 |
0.1260032017 |
- |
- |
PREDICTED: uncharacterized protein LOC104612097 isoform X2 [Nelumbo nucifera] |
| 14 |
Hb_003906_170 |
0.1268606745 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 15 |
Hb_069619_010 |
0.127817887 |
- |
- |
Aspartic proteinase precursor, putative [Ricinus communis] |
| 16 |
Hb_005081_020 |
0.1284162962 |
- |
- |
PREDICTED: ninja-family protein mc410 [Jatropha curcas] |
| 17 |
Hb_009732_020 |
0.1289682809 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Jatropha curcas] |
| 18 |
Hb_003626_050 |
0.1299671231 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 19 |
Hb_083799_010 |
0.1300924596 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 20 |
Hb_000157_070 |
0.1311703298 |
- |
- |
PREDICTED: alpha-N-acetylglucosaminidase [Jatropha curcas] |