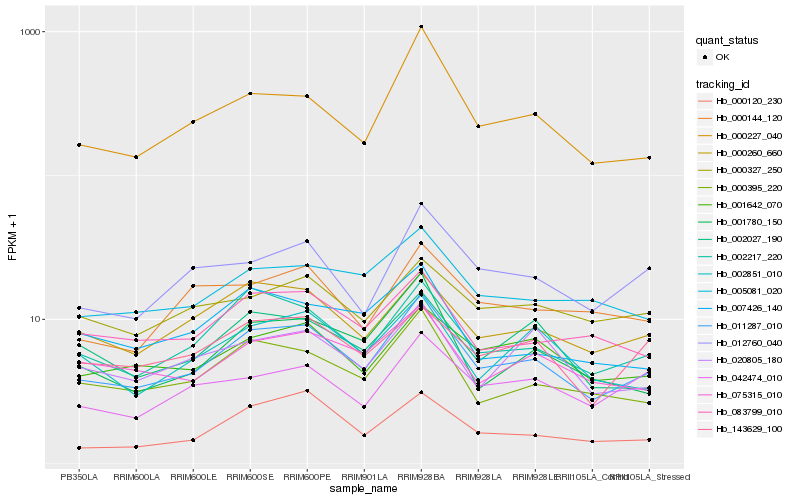
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011287_010 |
0.0 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 2 |
Hb_002217_220 |
0.081337699 |
- |
- |
hypothetical protein POPTR_0010s15930g [Populus trichocarpa] |
| 3 |
Hb_000395_220 |
0.0918729247 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 4 |
Hb_000227_040 |
0.0933439812 |
- |
- |
Stem-specific protein TSJT1, putative [Ricinus communis] |
| 5 |
Hb_000260_660 |
0.0933654123 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001642_070 |
0.1015876147 |
- |
- |
PREDICTED: uncharacterized protein LOC105646431 [Jatropha curcas] |
| 7 |
Hb_012760_040 |
0.1039093105 |
- |
- |
PREDICTED: aconitate hydratase 1 [Jatropha curcas] |
| 8 |
Hb_075315_010 |
0.1068283323 |
- |
- |
ornithine aminotransferase [Camellia sinensis] |
| 9 |
Hb_083799_010 |
0.108557648 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 10 |
Hb_001780_150 |
0.1091176865 |
- |
- |
ceramidase, putative [Ricinus communis] |
| 11 |
Hb_143629_100 |
0.1096753898 |
- |
- |
PREDICTED: phytochrome A [Jatropha curcas] |
| 12 |
Hb_005081_020 |
0.1098960709 |
- |
- |
PREDICTED: ninja-family protein mc410 [Jatropha curcas] |
| 13 |
Hb_002027_190 |
0.1130102622 |
- |
- |
PREDICTED: autophagy-related protein 18g [Jatropha curcas] |
| 14 |
Hb_002851_010 |
0.1147571247 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 15 |
Hb_000120_230 |
0.1165538868 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g33990-like [Jatropha curcas] |
| 16 |
Hb_020805_180 |
0.1190359841 |
- |
- |
PREDICTED: epidermal growth factor receptor substrate 15-like 1 [Jatropha curcas] |
| 17 |
Hb_042474_010 |
0.1191201638 |
- |
- |
PREDICTED: adenylyltransferase and sulfurtransferase MOCS3 [Jatropha curcas] |
| 18 |
Hb_007426_140 |
0.1194459367 |
- |
- |
ornithine aminotransferase, putative [Ricinus communis] |
| 19 |
Hb_000327_250 |
0.1205773102 |
- |
- |
PREDICTED: macrophage erythroblast attacher [Jatropha curcas] |
| 20 |
Hb_000144_120 |
0.1209659068 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |