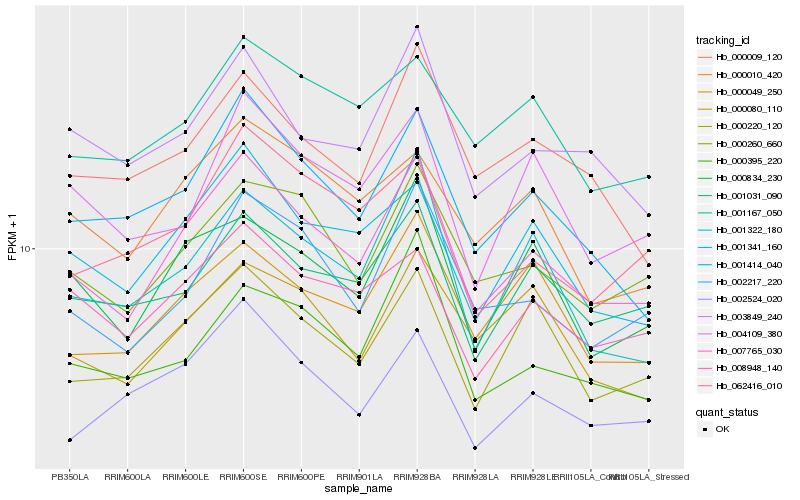
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007765_030 |
0.0 |
- |
- |
PREDICTED: neutral ceramidase [Jatropha curcas] |
| 2 |
Hb_002217_220 |
0.0793892006 |
- |
- |
hypothetical protein POPTR_0010s15930g [Populus trichocarpa] |
| 3 |
Hb_000260_660 |
0.0804209503 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001414_040 |
0.0814538002 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 1-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_001341_160 |
0.0829108853 |
- |
- |
PREDICTED: E3 SUMO-protein ligase SIZ1 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000220_120 |
0.0890435285 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000834_230 |
0.091442995 |
- |
- |
PREDICTED: ras-related protein RHN1-like [Jatropha curcas] |
| 8 |
Hb_003849_240 |
0.0925855815 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 9 |
Hb_001031_090 |
0.0930248914 |
- |
- |
PREDICTED: translational activator GCN1 [Jatropha curcas] |
| 10 |
Hb_008948_140 |
0.0962428782 |
- |
- |
PREDICTED: nuclear pore complex protein NUP205 [Jatropha curcas] |
| 11 |
Hb_000049_250 |
0.0967598574 |
- |
- |
PREDICTED: uncharacterized protein LOC105644455 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000010_420 |
0.098660847 |
- |
- |
Callose synthase 10 [Morus notabilis] |
| 13 |
Hb_000009_120 |
0.0993097078 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase P isoform X1 [Jatropha curcas] |
| 14 |
Hb_001322_180 |
0.0993468602 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 26 [Jatropha curcas] |
| 15 |
Hb_001167_050 |
0.1006077253 |
- |
- |
PREDICTED: uncharacterized protein LOC105648014 [Jatropha curcas] |
| 16 |
Hb_002524_020 |
0.1019964138 |
- |
- |
glycerophosphoryl diester phosphodiesterase, putative [Ricinus communis] |
| 17 |
Hb_062416_010 |
0.1024508811 |
- |
- |
PREDICTED: UPF0202 protein At1g10490 [Jatropha curcas] |
| 18 |
Hb_000395_220 |
0.1027579341 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 19 |
Hb_000080_110 |
0.1029542989 |
- |
- |
PREDICTED: GPI ethanolamine phosphate transferase 3 isoform X1 [Jatropha curcas] |
| 20 |
Hb_004109_380 |
0.1034376513 |
- |
- |
PREDICTED: callose synthase 9 isoform X4 [Pyrus x bretschneideri] |