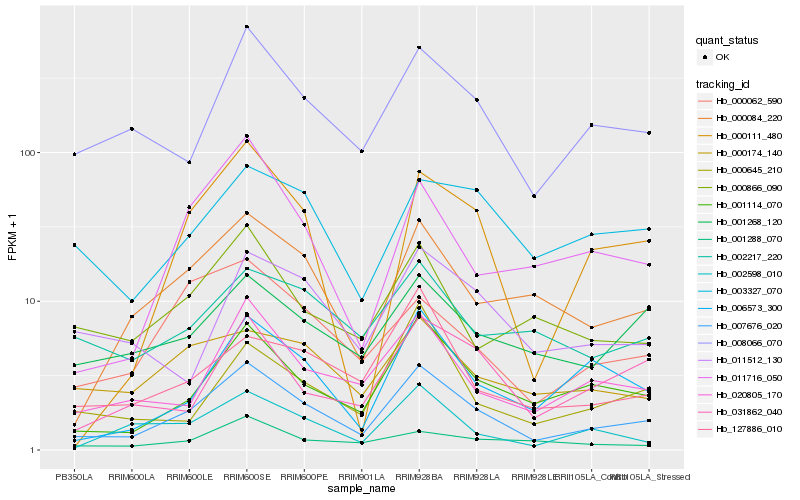
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007676_020 |
0.0 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase LIN-1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001114_070 |
0.1316276619 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At1g58400 [Jatropha curcas] |
| 3 |
Hb_008066_070 |
0.1473431659 |
transcription factor |
TF Family: ERF |
AP2/ERF super family protein [Hevea brasiliensis] |
| 4 |
Hb_020805_170 |
0.1545146127 |
- |
- |
PREDICTED: F-box protein CPR30-like [Jatropha curcas] |
| 5 |
Hb_000084_220 |
0.1661068068 |
- |
- |
PREDICTED: uncharacterized protein LOC105634692 [Jatropha curcas] |
| 6 |
Hb_006573_300 |
0.169081864 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 7 |
Hb_031862_040 |
0.1788325464 |
- |
- |
purine transporter, putative [Ricinus communis] |
| 8 |
Hb_001268_120 |
0.1790086894 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002598_010 |
0.1804358795 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 10 |
Hb_000174_140 |
0.1809358543 |
- |
- |
JHL23J11.8 [Jatropha curcas] |
| 11 |
Hb_000866_090 |
0.1820192262 |
- |
- |
PREDICTED: U-box domain-containing protein 33-like isoform X3 [Jatropha curcas] |
| 12 |
Hb_000062_590 |
0.1840018871 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 13 |
Hb_127886_010 |
0.1847132622 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 14 |
Hb_011716_050 |
0.1849102397 |
- |
- |
serine acetyltransferase [Hevea brasiliensis] |
| 15 |
Hb_001288_070 |
0.1853158273 |
- |
- |
PREDICTED: aminopeptidase M1-like [Jatropha curcas] |
| 16 |
Hb_000645_210 |
0.1875046363 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 5-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_000111_480 |
0.1877911335 |
- |
- |
PREDICTED: probable carboxylesterase 17 [Jatropha curcas] |
| 18 |
Hb_011512_130 |
0.1891983874 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At2g33170 [Jatropha curcas] |
| 19 |
Hb_002217_220 |
0.1898711921 |
- |
- |
hypothetical protein POPTR_0010s15930g [Populus trichocarpa] |
| 20 |
Hb_003327_070 |
0.1899082912 |
- |
- |
PREDICTED: UDP-glycosyltransferase 73B4-like [Jatropha curcas] |