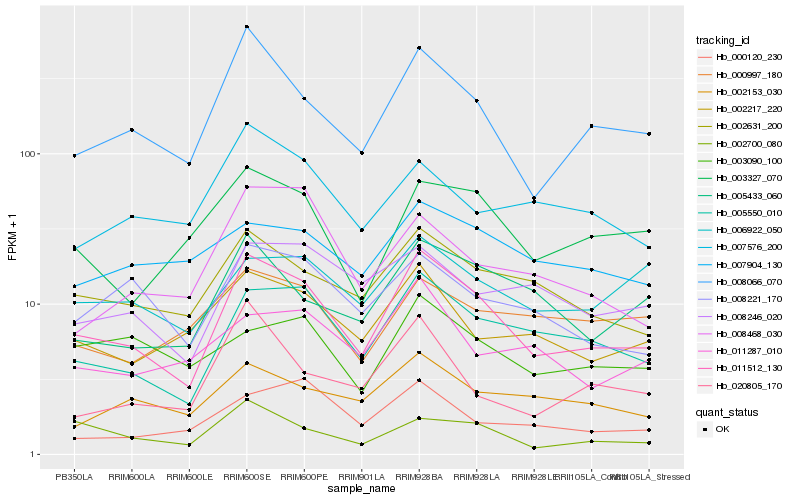
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011512_130 |
0.0 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At2g33170 [Jatropha curcas] |
| 2 |
Hb_005550_010 |
0.1151718924 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 4-like [Jatropha curcas] |
| 3 |
Hb_008066_070 |
0.1221784429 |
transcription factor |
TF Family: ERF |
AP2/ERF super family protein [Hevea brasiliensis] |
| 4 |
Hb_002631_200 |
0.1385899954 |
- |
- |
PREDICTED: uncharacterized protein LOC105646171 isoform X2 [Jatropha curcas] |
| 5 |
Hb_002217_220 |
0.1531516073 |
- |
- |
hypothetical protein POPTR_0010s15930g [Populus trichocarpa] |
| 6 |
Hb_005433_060 |
0.1564651554 |
- |
- |
PREDICTED: putative 1-phosphatidylinositol-3-phosphate 5-kinase FAB1C [Jatropha curcas] |
| 7 |
Hb_008221_170 |
0.1596080393 |
transcription factor |
TF Family: BES1 |
PREDICTED: BES1/BZR1 homolog protein 4 isoform X1 [Populus euphratica] |
| 8 |
Hb_003327_070 |
0.1597248286 |
- |
- |
PREDICTED: UDP-glycosyltransferase 73B4-like [Jatropha curcas] |
| 9 |
Hb_002700_080 |
0.16425339 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_24521 [Jatropha curcas] |
| 10 |
Hb_002153_030 |
0.1651003932 |
- |
- |
E3 ubiquitin-protein ligase UPL5 [Theobroma cacao] |
| 11 |
Hb_008246_020 |
0.1682617608 |
- |
- |
amidophosphoribosyltransferase, putative [Ricinus communis] |
| 12 |
Hb_000120_230 |
0.1700988498 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g33990-like [Jatropha curcas] |
| 13 |
Hb_011287_010 |
0.1716983746 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 14 |
Hb_007576_200 |
0.1740912913 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 15 |
Hb_006922_050 |
0.1758411986 |
- |
- |
PREDICTED: dipeptidyl peptidase 8 [Jatropha curcas] |
| 16 |
Hb_007904_130 |
0.1771784604 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 17 |
Hb_020805_170 |
0.1784182322 |
- |
- |
PREDICTED: F-box protein CPR30-like [Jatropha curcas] |
| 18 |
Hb_008468_030 |
0.1798010962 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 19 |
Hb_000997_180 |
0.1798322022 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003090_100 |
0.1799706933 |
- |
- |
PREDICTED: uncharacterized protein LOC105646670 [Jatropha curcas] |