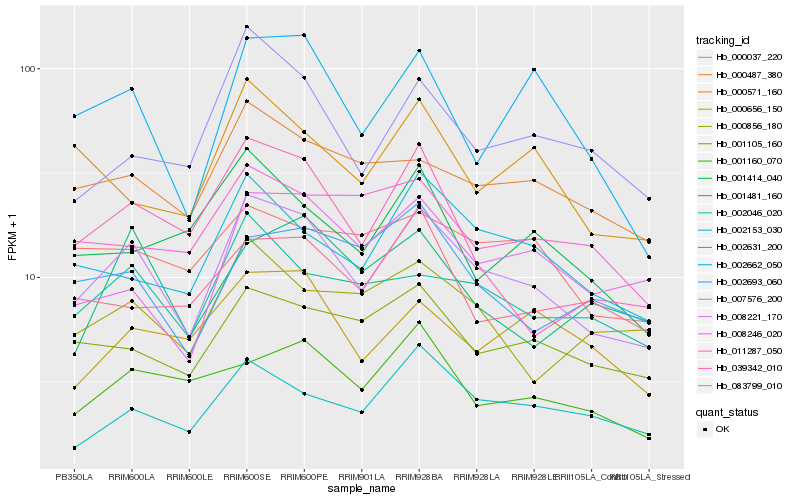
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008221_170 |
0.0 |
transcription factor |
TF Family: BES1 |
PREDICTED: BES1/BZR1 homolog protein 4 isoform X1 [Populus euphratica] |
| 2 |
Hb_002046_020 |
0.1252993899 |
- |
- |
PREDICTED: putative clathrin assembly protein At2g25430 [Jatropha curcas] |
| 3 |
Hb_002631_200 |
0.1257442923 |
- |
- |
PREDICTED: uncharacterized protein LOC105646171 isoform X2 [Jatropha curcas] |
| 4 |
Hb_001105_160 |
0.127991527 |
- |
- |
PREDICTED: protein phosphatase 2C 32 [Jatropha curcas] |
| 5 |
Hb_000487_380 |
0.1290384233 |
- |
- |
PREDICTED: protein PAT1 homolog 1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002662_050 |
0.1309918362 |
- |
- |
cysteine protease CP1 [Manihot esculenta] |
| 7 |
Hb_007576_200 |
0.1326815851 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 8 |
Hb_002153_030 |
0.1327705746 |
- |
- |
E3 ubiquitin-protein ligase UPL5 [Theobroma cacao] |
| 9 |
Hb_008246_020 |
0.1341448184 |
- |
- |
amidophosphoribosyltransferase, putative [Ricinus communis] |
| 10 |
Hb_039342_010 |
0.1358899023 |
- |
- |
pyruvate decarboxylase [Hevea brasiliensis] |
| 11 |
Hb_000571_160 |
0.1415082425 |
- |
- |
PREDICTED: ubiquitin-activating enzyme E1 1-like [Jatropha curcas] |
| 12 |
Hb_002693_060 |
0.1418204098 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 13 |
Hb_000656_150 |
0.1430400882 |
- |
- |
PREDICTED: exocyst complex component EXO70A1-like [Jatropha curcas] |
| 14 |
Hb_000856_180 |
0.1438628907 |
- |
- |
calcium-dependent protein kinase [Hevea brasiliensis] |
| 15 |
Hb_001160_070 |
0.1444222055 |
- |
- |
PREDICTED: uncharacterized protein LOC105632163 [Jatropha curcas] |
| 16 |
Hb_001414_040 |
0.1455543727 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 1-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_000037_220 |
0.146071855 |
- |
- |
PREDICTED: vacuolar protein-sorting-associated protein 11 homolog [Jatropha curcas] |
| 18 |
Hb_001481_160 |
0.1484310513 |
- |
- |
PREDICTED: RING-H2 finger protein ATL22-like [Populus euphratica] |
| 19 |
Hb_011287_050 |
0.1484744509 |
- |
- |
PREDICTED: uncharacterized protein LOC105644786 [Jatropha curcas] |
| 20 |
Hb_083799_010 |
0.1485351528 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |