| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
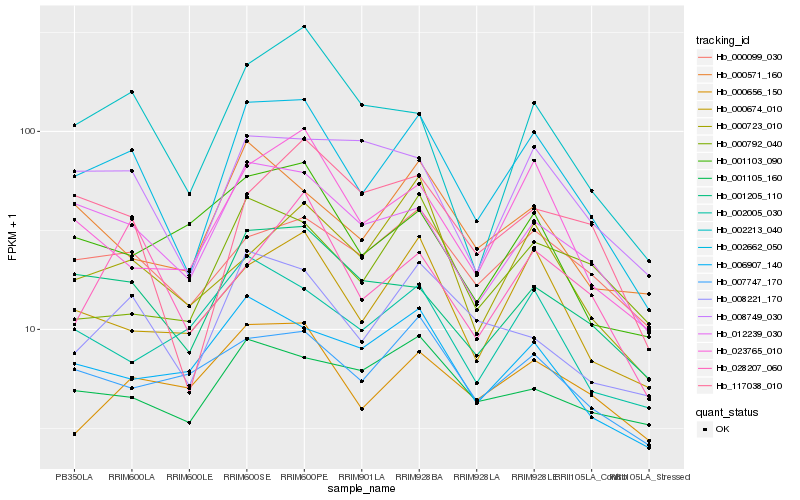
Hb_002662_050 |
0.0 |
- |
- |
cysteine protease CP1 [Manihot esculenta] |
| 2 |
Hb_012239_030 |
0.121560468 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 3 |
Hb_000674_010 |
0.1279840416 |
- |
- |
PREDICTED: copper-transporting ATPase RAN1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001205_110 |
0.1303976198 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 4 [Jatropha curcas] |
| 5 |
Hb_008221_170 |
0.1309918362 |
transcription factor |
TF Family: BES1 |
PREDICTED: BES1/BZR1 homolog protein 4 isoform X1 [Populus euphratica] |
| 6 |
Hb_023765_010 |
0.1341340332 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 7 |
Hb_000099_030 |
0.1397227482 |
- |
- |
6-phosphogluconate dehydrogenase, putative [Ricinus communis] |
| 8 |
Hb_008749_030 |
0.1445204719 |
- |
- |
hydroxyproline-rich glycoprotein [Populus trichocarpa] |
| 9 |
Hb_006907_140 |
0.1457481698 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000571_160 |
0.1474615283 |
- |
- |
PREDICTED: ubiquitin-activating enzyme E1 1-like [Jatropha curcas] |
| 11 |
Hb_117038_010 |
0.1498420827 |
- |
- |
cysteine protease [Hevea brasiliensis] |
| 12 |
Hb_002213_040 |
0.1498620478 |
- |
- |
cinnamoyl-CoA reductase [Hevea brasiliensis] |
| 13 |
Hb_000656_150 |
0.1500679018 |
- |
- |
PREDICTED: exocyst complex component EXO70A1-like [Jatropha curcas] |
| 14 |
Hb_007747_170 |
0.1518398333 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 [Jatropha curcas] |
| 15 |
Hb_000723_010 |
0.1526977425 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 16 |
Hb_002005_030 |
0.1586253795 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 17 |
Hb_028207_060 |
0.1598564872 |
- |
- |
PREDICTED: uncharacterized protein LOC105639235 [Jatropha curcas] |
| 18 |
Hb_001105_160 |
0.1607879758 |
- |
- |
PREDICTED: protein phosphatase 2C 32 [Jatropha curcas] |
| 19 |
Hb_000792_040 |
0.1610511244 |
- |
- |
PREDICTED: steroid 5-alpha-reductase DET2 [Jatropha curcas] |
| 20 |
Hb_001103_090 |
0.1619713273 |
- |
- |
PREDICTED: patellin-3 [Populus euphratica] |