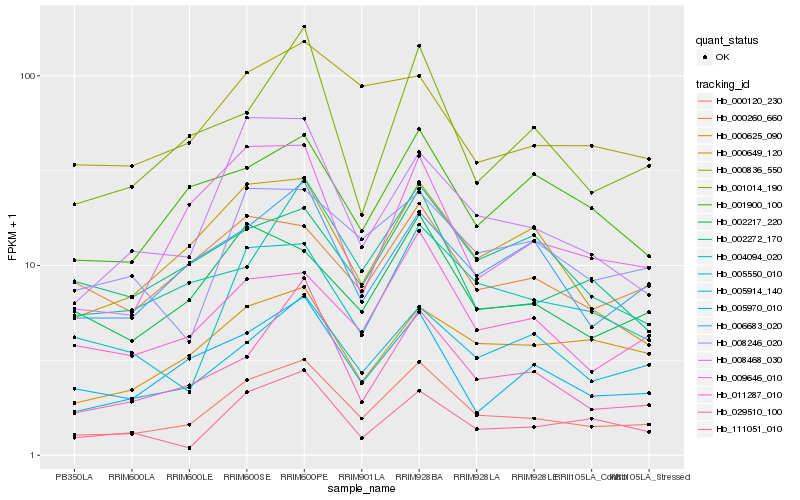
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000120_230 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g33990-like [Jatropha curcas] |
| 2 |
Hb_011287_010 |
0.1165538868 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 3 |
Hb_029510_100 |
0.1220659419 |
- |
- |
scab resistance family protein [Populus trichocarpa] |
| 4 |
Hb_008468_030 |
0.1239749763 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 5 |
Hb_006683_020 |
0.1256948988 |
- |
- |
PREDICTED: nucleosome assembly protein 1;4 [Jatropha curcas] |
| 6 |
Hb_005914_140 |
0.1266674197 |
- |
- |
PREDICTED: uncharacterized protein LOC105632183 [Jatropha curcas] |
| 7 |
Hb_005970_010 |
0.1266961328 |
- |
- |
PREDICTED: probable isoprenylcysteine alpha-carbonyl methylesterase ICMEL1 isoform X2 [Jatropha curcas] |
| 8 |
Hb_005550_010 |
0.1282645749 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 4-like [Jatropha curcas] |
| 9 |
Hb_000649_120 |
0.1287324178 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_002217_220 |
0.1288606601 |
- |
- |
hypothetical protein POPTR_0010s15930g [Populus trichocarpa] |
| 11 |
Hb_001014_190 |
0.1316539673 |
- |
- |
PREDICTED: uncharacterized protein LOC105647294 [Jatropha curcas] |
| 12 |
Hb_009646_010 |
0.1365038026 |
- |
- |
PREDICTED: hyoscyamine 6-dioxygenase-like [Jatropha curcas] |
| 13 |
Hb_111051_010 |
0.1370201841 |
- |
- |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 14 |
Hb_000625_090 |
0.1382824684 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_008246_020 |
0.1404218755 |
- |
- |
amidophosphoribosyltransferase, putative [Ricinus communis] |
| 16 |
Hb_002272_170 |
0.1410287817 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase-like protein 1 [Jatropha curcas] |
| 17 |
Hb_001900_100 |
0.1413138257 |
- |
- |
PREDICTED: uncharacterized protein LOC105632664 isoform X1 [Jatropha curcas] |
| 18 |
Hb_004094_020 |
0.142996942 |
- |
- |
Pyruvate kinase family protein isoform 1 [Theobroma cacao] |
| 19 |
Hb_000836_550 |
0.1435688641 |
- |
- |
PREDICTED: reticulon-like protein B5 [Jatropha curcas] |
| 20 |
Hb_000260_660 |
0.1442260975 |
- |
- |
conserved hypothetical protein [Ricinus communis] |