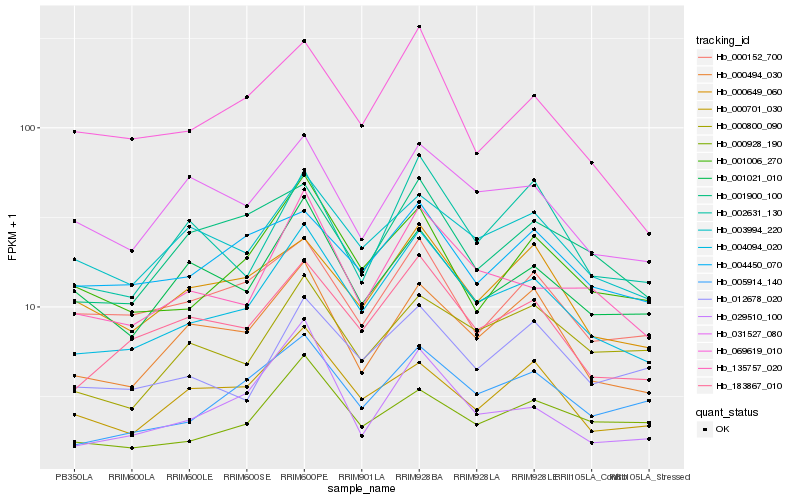
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004094_020 |
0.0 |
- |
- |
Pyruvate kinase family protein isoform 1 [Theobroma cacao] |
| 2 |
Hb_183867_010 |
0.0823959876 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62930, chloroplastic-like [Populus euphratica] |
| 3 |
Hb_135757_020 |
0.090775 |
- |
- |
hypothetical protein AMTR_s00420p00013340 [Amborella trichopoda] |
| 4 |
Hb_005914_140 |
0.1033485143 |
- |
- |
PREDICTED: uncharacterized protein LOC105632183 [Jatropha curcas] |
| 5 |
Hb_000494_030 |
0.1039611531 |
- |
- |
PREDICTED: uncharacterized protein LOC105643196 [Jatropha curcas] |
| 6 |
Hb_001006_270 |
0.1071573233 |
- |
- |
peptide transporter, putative [Ricinus communis] |
| 7 |
Hb_003994_220 |
0.1075585954 |
- |
- |
PREDICTED: V-type proton ATPase catalytic subunit A [Jatropha curcas] |
| 8 |
Hb_069619_010 |
0.1085015257 |
- |
- |
Aspartic proteinase precursor, putative [Ricinus communis] |
| 9 |
Hb_031527_080 |
0.1168027053 |
- |
- |
utp-glucose-1-phosphate uridylyltransferase, putative [Ricinus communis] |
| 10 |
Hb_000649_060 |
0.1174863444 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase RIE1 [Jatropha curcas] |
| 11 |
Hb_000701_030 |
0.1176657179 |
- |
- |
Sodium/hydrogen exchanger 6 -like protein [Gossypium arboreum] |
| 12 |
Hb_012678_020 |
0.1179100823 |
- |
- |
PREDICTED: outer envelope protein 61 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001900_100 |
0.1181557355 |
- |
- |
PREDICTED: uncharacterized protein LOC105632664 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001021_010 |
0.1194730097 |
- |
- |
PREDICTED: bifunctional protein FolD 2 [Jatropha curcas] |
| 15 |
Hb_000928_190 |
0.1209738084 |
- |
- |
PREDICTED: putative acyl-activating enzyme 19 isoform X1 [Jatropha curcas] |
| 16 |
Hb_002631_130 |
0.1211257229 |
- |
- |
PREDICTED: L-ascorbate oxidase-like [Jatropha curcas] |
| 17 |
Hb_029510_100 |
0.1214841766 |
- |
- |
scab resistance family protein [Populus trichocarpa] |
| 18 |
Hb_000800_090 |
0.1215427348 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 1 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 1 of pyruvate dehydrogenase complex, mitochondrial [Jatropha curcas] |
| 19 |
Hb_004450_070 |
0.1228094651 |
- |
- |
PREDICTED: 2-hydroxyacyl-CoA lyase [Jatropha curcas] |
| 20 |
Hb_000152_700 |
0.1230500241 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase RIN2 [Jatropha curcas] |