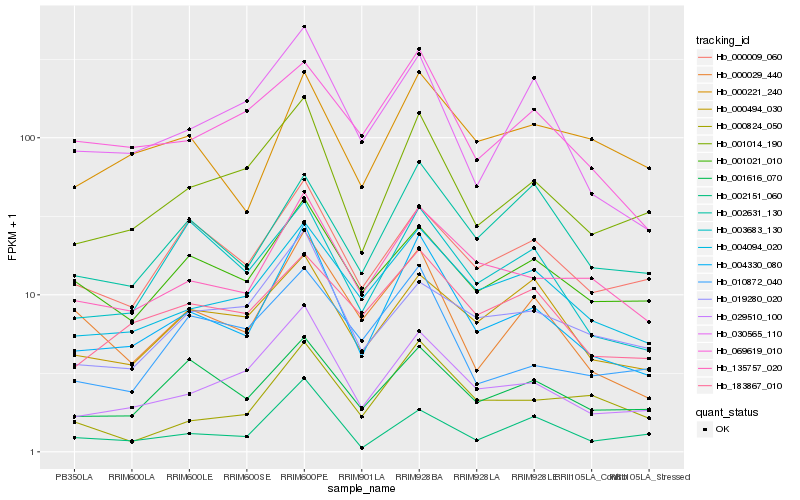
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004330_080 |
0.0 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 8 isoform X1 [Jatropha curcas] |
| 2 |
Hb_029510_100 |
0.1063321943 |
- |
- |
scab resistance family protein [Populus trichocarpa] |
| 3 |
Hb_001014_190 |
0.1110261326 |
- |
- |
PREDICTED: uncharacterized protein LOC105647294 [Jatropha curcas] |
| 4 |
Hb_135757_020 |
0.1194544433 |
- |
- |
hypothetical protein AMTR_s00420p00013340 [Amborella trichopoda] |
| 5 |
Hb_004094_020 |
0.1304560905 |
- |
- |
Pyruvate kinase family protein isoform 1 [Theobroma cacao] |
| 6 |
Hb_000029_440 |
0.1335937224 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase 48 kDa subunit isoform X2 [Vitis vinifera] |
| 7 |
Hb_030565_110 |
0.1459783293 |
- |
- |
PREDICTED: adenosylhomocysteinase 1 [Jatropha curcas] |
| 8 |
Hb_001616_070 |
0.1463694561 |
- |
- |
PREDICTED: uncharacterized protein LOC105644365 [Jatropha curcas] |
| 9 |
Hb_069619_010 |
0.1511998666 |
- |
- |
Aspartic proteinase precursor, putative [Ricinus communis] |
| 10 |
Hb_003683_130 |
0.1531283857 |
- |
- |
PREDICTED: enolase 1, chloroplastic [Jatropha curcas] |
| 11 |
Hb_183867_010 |
0.154563 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62930, chloroplastic-like [Populus euphratica] |
| 12 |
Hb_000009_060 |
0.1557470602 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 1B [Jatropha curcas] |
| 13 |
Hb_001021_010 |
0.1559602683 |
- |
- |
PREDICTED: bifunctional protein FolD 2 [Jatropha curcas] |
| 14 |
Hb_010872_040 |
0.1569485882 |
- |
- |
hypothetical protein F775_31993 [Aegilops tauschii] |
| 15 |
Hb_000221_240 |
0.157504525 |
- |
- |
chloroplast 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase [Hevea brasiliensis] |
| 16 |
Hb_000824_050 |
0.1580336193 |
- |
- |
PREDICTED: nicotinate phosphoribosyltransferase 1 [Jatropha curcas] |
| 17 |
Hb_019280_020 |
0.1611657107 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK14 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002631_130 |
0.1616170762 |
- |
- |
PREDICTED: L-ascorbate oxidase-like [Jatropha curcas] |
| 19 |
Hb_000494_030 |
0.1632427509 |
- |
- |
PREDICTED: uncharacterized protein LOC105643196 [Jatropha curcas] |
| 20 |
Hb_002151_060 |
0.1644314203 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit rpc4 [Jatropha curcas] |