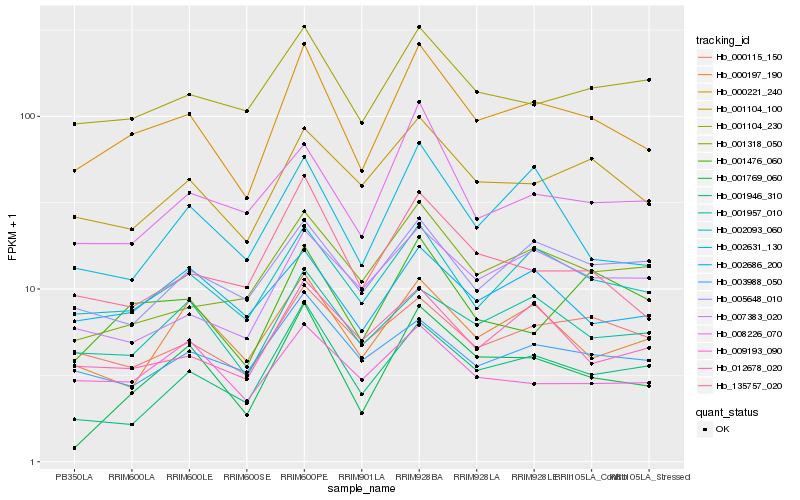
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000221_240 |
0.0 |
- |
- |
chloroplast 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase [Hevea brasiliensis] |
| 2 |
Hb_002093_060 |
0.1012154878 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 3 |
Hb_001104_100 |
0.1128029999 |
- |
- |
RecName: Full=Enolase 2; AltName: Full=2-phospho-D-glycerate hydro-lyase 2; AltName: Full=2-phosphoglycerate dehydratase 2; AltName: Allergen=Hev b 9 [Hevea brasiliensis] |
| 4 |
Hb_135757_020 |
0.1135181715 |
- |
- |
hypothetical protein AMTR_s00420p00013340 [Amborella trichopoda] |
| 5 |
Hb_001957_010 |
0.1195549113 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g14410 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001104_230 |
0.1208737657 |
- |
- |
PREDICTED: fructose-bisphosphate aldolase cytoplasmic isozyme [Jatropha curcas] |
| 7 |
Hb_001769_060 |
0.1212172238 |
- |
- |
PREDICTED: F-box protein At3g12350 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001476_060 |
0.1254294933 |
- |
- |
PREDICTED: uncharacterized protein At5g01610 [Jatropha curcas] |
| 9 |
Hb_001946_310 |
0.1255632963 |
- |
- |
PREDICTED: probable ethanolamine kinase isoform X2 [Jatropha curcas] |
| 10 |
Hb_012678_020 |
0.1261014322 |
- |
- |
PREDICTED: outer envelope protein 61 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002631_130 |
0.1266102589 |
- |
- |
PREDICTED: L-ascorbate oxidase-like [Jatropha curcas] |
| 12 |
Hb_002686_200 |
0.1301209385 |
- |
- |
glucosidase II beta subunit, putative [Ricinus communis] |
| 13 |
Hb_008226_070 |
0.1302471436 |
- |
- |
PREDICTED: expansin-A8 [Jatropha curcas] |
| 14 |
Hb_000115_150 |
0.1320351636 |
- |
- |
PREDICTED: IST1 homolog [Jatropha curcas] |
| 15 |
Hb_000197_190 |
0.1325238074 |
- |
- |
PREDICTED: diaminopimelate decarboxylase 2, chloroplastic-like [Jatropha curcas] |
| 16 |
Hb_003988_050 |
0.1327644297 |
- |
- |
PREDICTED: zinc finger protein-like 1 homolog [Jatropha curcas] |
| 17 |
Hb_001318_050 |
0.1345045765 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 18 |
Hb_005648_010 |
0.1352213462 |
- |
- |
PREDICTED: malate dehydrogenase, glyoxysomal isoform X1 [Jatropha curcas] |
| 19 |
Hb_009193_090 |
0.135544873 |
- |
- |
PREDICTED: protein root UVB sensitive 3 isoform X1 [Jatropha curcas] |
| 20 |
Hb_007383_020 |
0.1388999686 |
- |
- |
conserved hypothetical protein [Ricinus communis] |