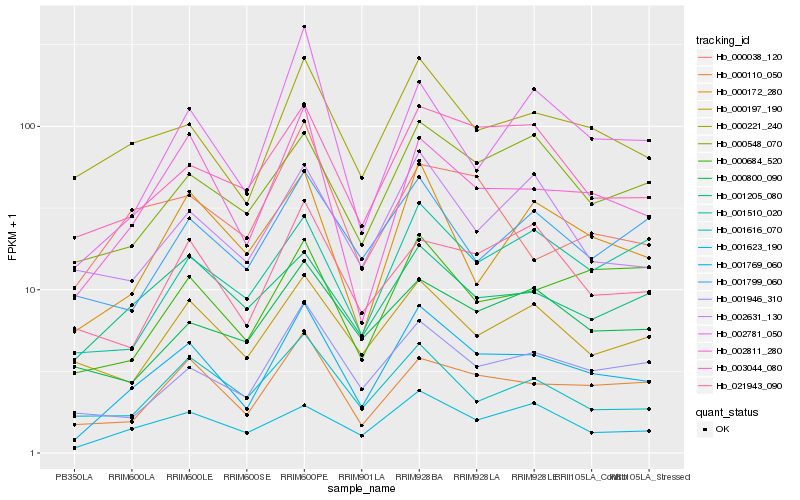
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001769_060 |
0.0 |
- |
- |
PREDICTED: F-box protein At3g12350 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002811_280 |
0.1176401068 |
- |
- |
PREDICTED: probable aldo-keto reductase 1 [Jatropha curcas] |
| 3 |
Hb_000221_240 |
0.1212172238 |
- |
- |
chloroplast 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase [Hevea brasiliensis] |
| 4 |
Hb_000110_050 |
0.1314620609 |
- |
- |
- |
| 5 |
Hb_001623_190 |
0.1334604299 |
- |
- |
PREDICTED: uncharacterized protein LOC105638473 [Jatropha curcas] |
| 6 |
Hb_001946_310 |
0.1373096467 |
- |
- |
PREDICTED: probable ethanolamine kinase isoform X2 [Jatropha curcas] |
| 7 |
Hb_003044_080 |
0.1472177538 |
desease resistance |
Gene Name: Synthase_beta |
RecName: Full=ATP synthase subunit beta, mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 8 |
Hb_000548_070 |
0.1494476751 |
desease resistance |
Gene Name: Synthase_beta |
RecName: Full=ATP synthase subunit beta, mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 9 |
Hb_000172_280 |
0.1510328568 |
- |
- |
aspartate aminotransferase, putative [Ricinus communis] |
| 10 |
Hb_001616_070 |
0.1513957257 |
- |
- |
PREDICTED: uncharacterized protein LOC105644365 [Jatropha curcas] |
| 11 |
Hb_000684_520 |
0.1565971507 |
- |
- |
glutathione S-transferase L3-like [Jatropha curcas] |
| 12 |
Hb_001205_080 |
0.1573145608 |
- |
- |
Protein FAM96B, putative [Ricinus communis] |
| 13 |
Hb_000197_190 |
0.1573950647 |
- |
- |
PREDICTED: diaminopimelate decarboxylase 2, chloroplastic-like [Jatropha curcas] |
| 14 |
Hb_002631_130 |
0.1604402638 |
- |
- |
PREDICTED: L-ascorbate oxidase-like [Jatropha curcas] |
| 15 |
Hb_001510_020 |
0.1607886285 |
- |
- |
Diaminopimelate epimerase, putative [Ricinus communis] |
| 16 |
Hb_021943_090 |
0.1654441743 |
- |
- |
PREDICTED: probable methylenetetrahydrofolate reductase [Jatropha curcas] |
| 17 |
Hb_001799_060 |
0.1660285421 |
- |
- |
Rab6 [Hevea brasiliensis] |
| 18 |
Hb_002781_050 |
0.1660473338 |
- |
- |
cytochrome B5 isoform 1, putative [Ricinus communis] |
| 19 |
Hb_000038_120 |
0.169444781 |
- |
- |
PREDICTED: endoglucanase 9-like [Jatropha curcas] |
| 20 |
Hb_000800_090 |
0.1698460195 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 1 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 1 of pyruvate dehydrogenase complex, mitochondrial [Jatropha curcas] |