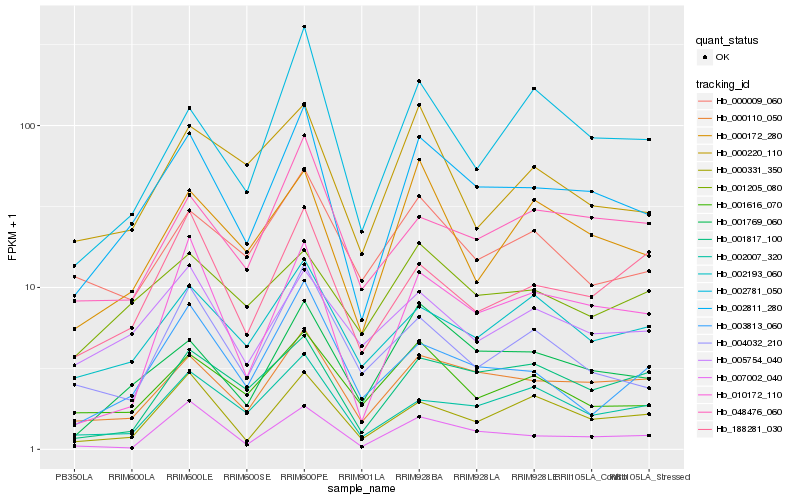
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002811_280 |
0.0 |
- |
- |
PREDICTED: probable aldo-keto reductase 1 [Jatropha curcas] |
| 2 |
Hb_000110_050 |
0.1051513108 |
- |
- |
- |
| 3 |
Hb_001769_060 |
0.1176401068 |
- |
- |
PREDICTED: F-box protein At3g12350 isoform X1 [Jatropha curcas] |
| 4 |
Hb_010172_110 |
0.1436452366 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 5 |
Hb_000172_280 |
0.1473081123 |
- |
- |
aspartate aminotransferase, putative [Ricinus communis] |
| 6 |
Hb_002007_320 |
0.1474242522 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase ANP1 [Jatropha curcas] |
| 7 |
Hb_001616_070 |
0.1492435023 |
- |
- |
PREDICTED: uncharacterized protein LOC105644365 [Jatropha curcas] |
| 8 |
Hb_002193_060 |
0.1494294883 |
- |
- |
PREDICTED: uncharacterized protein C4orf29 homolog [Pyrus x bretschneideri] |
| 9 |
Hb_000331_350 |
0.1522795003 |
- |
- |
hypothetical protein JCGZ_11337 [Jatropha curcas] |
| 10 |
Hb_048476_060 |
0.1523533144 |
- |
- |
PREDICTED: CRAL-TRIO domain-containing protein YKL091C-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_001817_100 |
0.1567580301 |
- |
- |
PREDICTED: deoxyhypusine hydroxylase [Jatropha curcas] |
| 12 |
Hb_002781_050 |
0.1574405469 |
- |
- |
cytochrome B5 isoform 1, putative [Ricinus communis] |
| 13 |
Hb_188281_030 |
0.1579323666 |
- |
- |
PREDICTED: uridine nucleosidase 1 [Jatropha curcas] |
| 14 |
Hb_005754_040 |
0.1586345143 |
- |
- |
PREDICTED: uncharacterized protein LOC105636678 isoform X2 [Jatropha curcas] |
| 15 |
Hb_004032_210 |
0.1609101387 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000220_110 |
0.1620346874 |
- |
- |
fructose-bisphosphate aldolase, putative [Ricinus communis] |
| 17 |
Hb_007002_040 |
0.1634885639 |
transcription factor |
TF Family: PHD |
DNA replication regulator dpb11, putative [Ricinus communis] |
| 18 |
Hb_000009_060 |
0.1636193908 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 1B [Jatropha curcas] |
| 19 |
Hb_001205_080 |
0.1641611631 |
- |
- |
Protein FAM96B, putative [Ricinus communis] |
| 20 |
Hb_003813_060 |
0.1651379547 |
- |
- |
conserved hypothetical protein [Ricinus communis] |