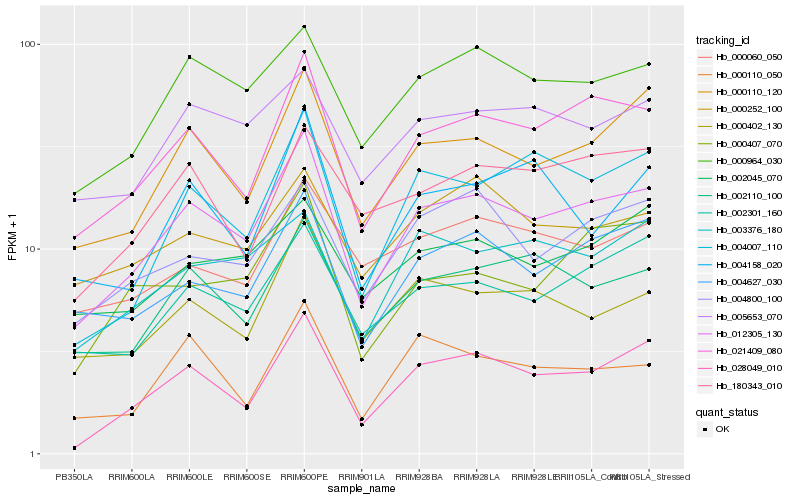
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012305_130 |
0.0 |
- |
- |
PREDICTED: GTP-binding protein SAR1A [Jatropha curcas] |
| 2 |
Hb_021409_080 |
0.0646899105 |
- |
- |
PREDICTED: ORM1-like protein 1 [Jatropha curcas] |
| 3 |
Hb_000110_120 |
0.0835674353 |
- |
- |
PREDICTED: ras-related protein RABE1a [Fragaria vesca subsp. vesca] |
| 4 |
Hb_000964_030 |
0.0911908493 |
- |
- |
ADP/ATP carrier 2 [Theobroma cacao] |
| 5 |
Hb_028049_010 |
0.0919426333 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002110_100 |
0.0932940556 |
- |
- |
PREDICTED: pyridoxine/pyridoxamine 5'-phosphate oxidase 1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_004627_030 |
0.09444785 |
- |
- |
ubiquitin-conjugating enzyme E2 C, putative [Ricinus communis] |
| 8 |
Hb_002301_160 |
0.0953255347 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g04780-like [Jatropha curcas] |
| 9 |
Hb_000402_130 |
0.1033577596 |
- |
- |
PREDICTED: AP-1 complex subunit mu-2 [Jatropha curcas] |
| 10 |
Hb_005653_070 |
0.1065853813 |
- |
- |
K+ transport growth defect-like protein [Hevea brasiliensis] |
| 11 |
Hb_004007_110 |
0.1077227445 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin 2-like [Jatropha curcas] |
| 12 |
Hb_004800_100 |
0.1088696014 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000407_070 |
0.1125653436 |
- |
- |
PREDICTED: probable tyrosine-protein phosphatase At1g05000 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000110_050 |
0.1134615019 |
- |
- |
- |
| 15 |
Hb_003376_180 |
0.113655569 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase MBR1 [Jatropha curcas] |
| 16 |
Hb_002045_070 |
0.1139381355 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor UNE12-like [Jatropha curcas] |
| 17 |
Hb_000252_100 |
0.1170880571 |
- |
- |
PREDICTED: AP-1 complex subunit mu-2 [Jatropha curcas] |
| 18 |
Hb_180343_010 |
0.1171480226 |
- |
- |
Biotin carboxyl carrier protein of acetyl-CoA carboxylase, putative [Ricinus communis] |
| 19 |
Hb_000060_050 |
0.1174426356 |
- |
- |
PREDICTED: uncharacterized protein LOC105649718 [Jatropha curcas] |
| 20 |
Hb_004158_020 |
0.1177569976 |
- |
- |
Rab1 [Hevea brasiliensis] |