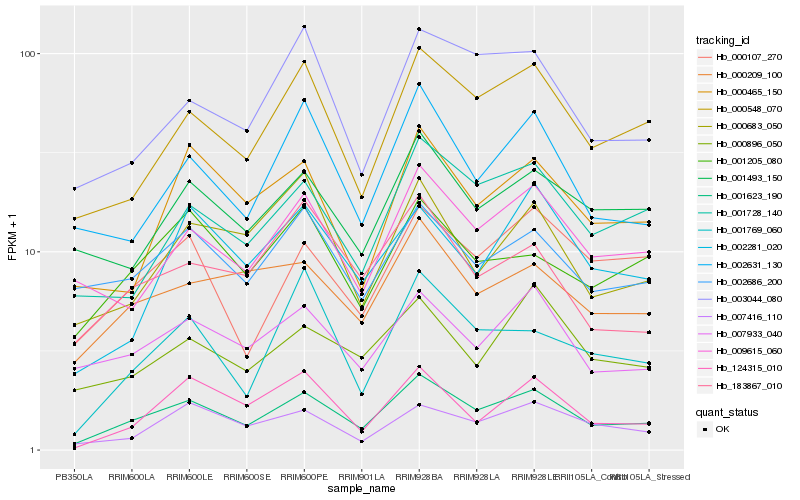
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001623_190 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638473 [Jatropha curcas] |
| 2 |
Hb_000548_070 |
0.103466577 |
desease resistance |
Gene Name: Synthase_beta |
RecName: Full=ATP synthase subunit beta, mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 3 |
Hb_002631_130 |
0.1186876405 |
- |
- |
PREDICTED: L-ascorbate oxidase-like [Jatropha curcas] |
| 4 |
Hb_001728_140 |
0.1196374241 |
- |
- |
- |
| 5 |
Hb_007933_040 |
0.1216467066 |
- |
- |
PREDICTED: probable beta-D-xylosidase 6 isoform X2 [Jatropha curcas] |
| 6 |
Hb_009615_060 |
0.1218580812 |
- |
- |
PREDICTED: prolyl endopeptidase-like [Jatropha curcas] |
| 7 |
Hb_003044_080 |
0.1229887867 |
desease resistance |
Gene Name: Synthase_beta |
RecName: Full=ATP synthase subunit beta, mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 8 |
Hb_000209_100 |
0.1238525124 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 2 isoform X2 [Jatropha curcas] |
| 9 |
Hb_007416_110 |
0.1269467426 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 14 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001205_080 |
0.1271173184 |
- |
- |
Protein FAM96B, putative [Ricinus communis] |
| 11 |
Hb_000465_150 |
0.1275782299 |
- |
- |
PREDICTED: peroxisomal acyl-coenzyme A oxidase 1 [Jatropha curcas] |
| 12 |
Hb_002686_200 |
0.1286085146 |
- |
- |
glucosidase II beta subunit, putative [Ricinus communis] |
| 13 |
Hb_000107_270 |
0.1297963199 |
- |
- |
Histone deacetylase 1 isoform 1 [Theobroma cacao] |
| 14 |
Hb_001493_150 |
0.1307438309 |
- |
- |
PREDICTED: alanine aminotransferase 2-like [Jatropha curcas] |
| 15 |
Hb_000683_050 |
0.1330628265 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 16 |
Hb_001769_060 |
0.1334604299 |
- |
- |
PREDICTED: F-box protein At3g12350 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000896_050 |
0.1337584309 |
- |
- |
PREDICTED: uncharacterized protein LOC105637668 [Jatropha curcas] |
| 18 |
Hb_183867_010 |
0.1338282715 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62930, chloroplastic-like [Populus euphratica] |
| 19 |
Hb_002281_020 |
0.1352203357 |
- |
- |
PREDICTED: actin-related protein 8 [Jatropha curcas] |
| 20 |
Hb_124315_010 |
0.1352930678 |
- |
- |
PREDICTED: uncharacterized protein LOC105650413 [Jatropha curcas] |