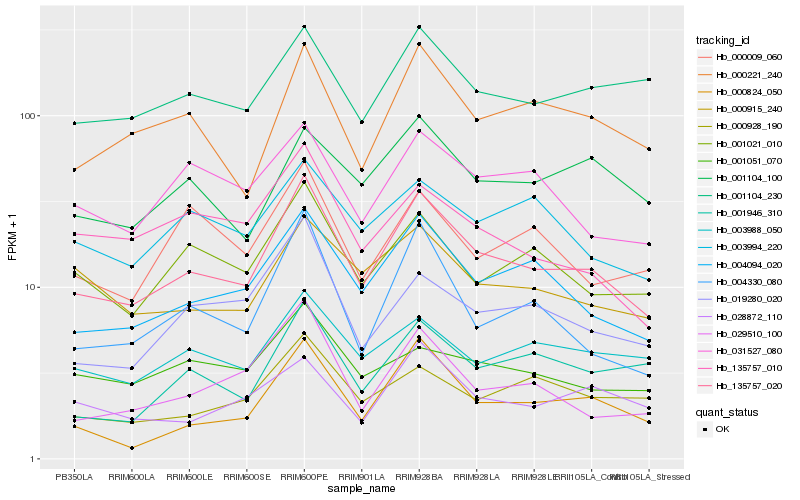
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_135757_020 |
0.0 |
- |
- |
hypothetical protein AMTR_s00420p00013340 [Amborella trichopoda] |
| 2 |
Hb_004094_020 |
0.090775 |
- |
- |
Pyruvate kinase family protein isoform 1 [Theobroma cacao] |
| 3 |
Hb_000221_240 |
0.1135181715 |
- |
- |
chloroplast 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase [Hevea brasiliensis] |
| 4 |
Hb_001021_010 |
0.117658197 |
- |
- |
PREDICTED: bifunctional protein FolD 2 [Jatropha curcas] |
| 5 |
Hb_004330_080 |
0.1194544433 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 8 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000824_050 |
0.1238424627 |
- |
- |
PREDICTED: nicotinate phosphoribosyltransferase 1 [Jatropha curcas] |
| 7 |
Hb_019280_020 |
0.1245386516 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK14 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000915_240 |
0.1256308469 |
- |
- |
catalytic, putative [Ricinus communis] |
| 9 |
Hb_031527_080 |
0.1276112899 |
- |
- |
utp-glucose-1-phosphate uridylyltransferase, putative [Ricinus communis] |
| 10 |
Hb_003988_050 |
0.1289746338 |
- |
- |
PREDICTED: zinc finger protein-like 1 homolog [Jatropha curcas] |
| 11 |
Hb_029510_100 |
0.1291855609 |
- |
- |
scab resistance family protein [Populus trichocarpa] |
| 12 |
Hb_001104_230 |
0.1302778685 |
- |
- |
PREDICTED: fructose-bisphosphate aldolase cytoplasmic isozyme [Jatropha curcas] |
| 13 |
Hb_003994_220 |
0.1320105146 |
- |
- |
PREDICTED: V-type proton ATPase catalytic subunit A [Jatropha curcas] |
| 14 |
Hb_001104_100 |
0.1347872321 |
- |
- |
RecName: Full=Enolase 2; AltName: Full=2-phospho-D-glycerate hydro-lyase 2; AltName: Full=2-phosphoglycerate dehydratase 2; AltName: Allergen=Hev b 9 [Hevea brasiliensis] |
| 15 |
Hb_000928_190 |
0.1357195862 |
- |
- |
PREDICTED: putative acyl-activating enzyme 19 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001946_310 |
0.1359021727 |
- |
- |
PREDICTED: probable ethanolamine kinase isoform X2 [Jatropha curcas] |
| 17 |
Hb_028872_110 |
0.1359994017 |
- |
- |
PREDICTED: uncharacterized protein LOC105644776 [Jatropha curcas] |
| 18 |
Hb_001051_070 |
0.1361427349 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000009_060 |
0.1364029829 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 1B [Jatropha curcas] |
| 20 |
Hb_135757_010 |
0.1370085553 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.6 [Jatropha curcas] |