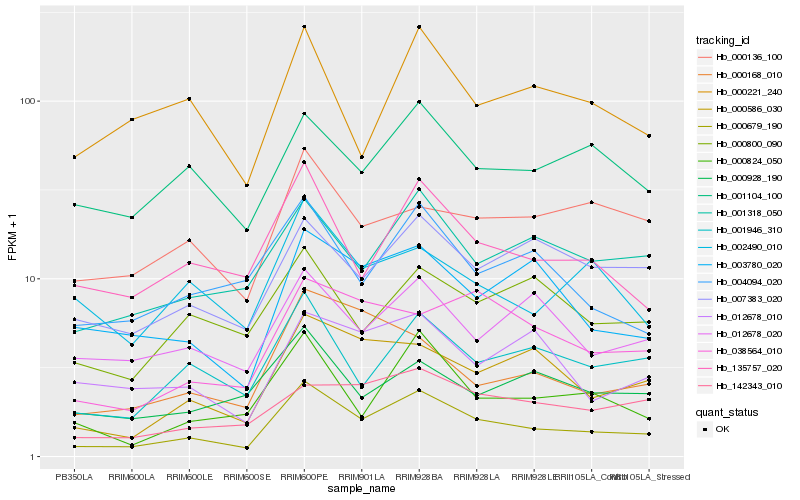
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000679_190 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105637641 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001946_310 |
0.145138632 |
- |
- |
PREDICTED: probable ethanolamine kinase isoform X2 [Jatropha curcas] |
| 3 |
Hb_000824_050 |
0.146131384 |
- |
- |
PREDICTED: nicotinate phosphoribosyltransferase 1 [Jatropha curcas] |
| 4 |
Hb_135757_020 |
0.1469490576 |
- |
- |
hypothetical protein AMTR_s00420p00013340 [Amborella trichopoda] |
| 5 |
Hb_000586_030 |
0.1497980805 |
- |
- |
hypothetical protein OsJ_17572 [Oryza sativa Japonica Group] |
| 6 |
Hb_001318_050 |
0.154604398 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 7 |
Hb_003780_020 |
0.1552150883 |
- |
- |
PREDICTED: prolyl 4-hydroxylase subunit alpha-1 isoform X2 [Glycine max] |
| 8 |
Hb_004094_020 |
0.1627966126 |
- |
- |
Pyruvate kinase family protein isoform 1 [Theobroma cacao] |
| 9 |
Hb_007383_020 |
0.1650263996 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_038564_010 |
0.1681846474 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 5 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000168_010 |
0.1711035531 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RFWD3-like isoform X1 [Populus euphratica] |
| 12 |
Hb_001104_100 |
0.172201931 |
- |
- |
RecName: Full=Enolase 2; AltName: Full=2-phospho-D-glycerate hydro-lyase 2; AltName: Full=2-phosphoglycerate dehydratase 2; AltName: Allergen=Hev b 9 [Hevea brasiliensis] |
| 13 |
Hb_142343_010 |
0.1724807156 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 14 |
Hb_012678_020 |
0.17251265 |
- |
- |
PREDICTED: outer envelope protein 61 isoform X1 [Jatropha curcas] |
| 15 |
Hb_012678_010 |
0.1739248064 |
- |
- |
PREDICTED: outer envelope protein 61-like [Gossypium raimondii] |
| 16 |
Hb_000221_240 |
0.1761033056 |
- |
- |
chloroplast 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase [Hevea brasiliensis] |
| 17 |
Hb_000800_090 |
0.1775149697 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 1 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 1 of pyruvate dehydrogenase complex, mitochondrial [Jatropha curcas] |
| 18 |
Hb_002490_010 |
0.1777075804 |
- |
- |
PREDICTED: plastidic glucose transporter 4 [Jatropha curcas] |
| 19 |
Hb_000928_190 |
0.1798130241 |
- |
- |
PREDICTED: putative acyl-activating enzyme 19 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000136_100 |
0.1813867832 |
- |
- |
PREDICTED: syntaxin-61 [Jatropha curcas] |