| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
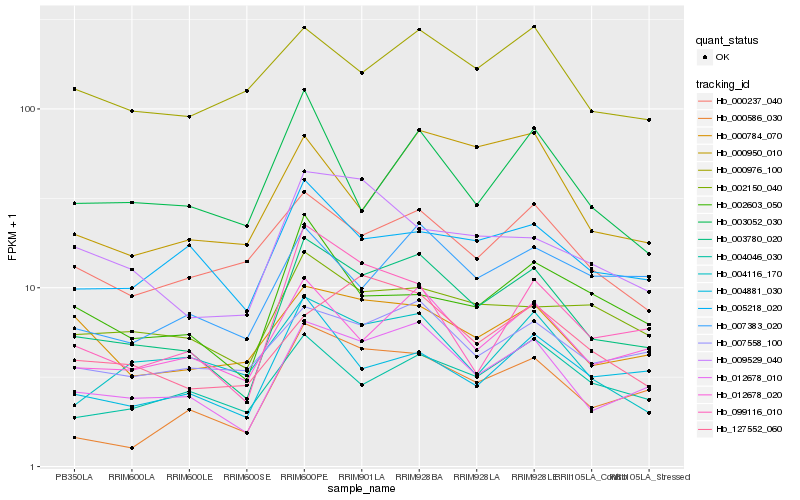
Hb_003780_020 |
0.0 |
- |
- |
PREDICTED: prolyl 4-hydroxylase subunit alpha-1 isoform X2 [Glycine max] |
| 2 |
Hb_012678_010 |
0.0582941461 |
- |
- |
PREDICTED: outer envelope protein 61-like [Gossypium raimondii] |
| 3 |
Hb_012678_020 |
0.1103267034 |
- |
- |
PREDICTED: outer envelope protein 61 isoform X1 [Jatropha curcas] |
| 4 |
Hb_007558_100 |
0.1328404217 |
- |
- |
PREDICTED: uncharacterized protein LOC105649092 isoform X2 [Jatropha curcas] |
| 5 |
Hb_002150_040 |
0.1368924249 |
- |
- |
PREDICTED: CMP-sialic acid transporter 1 isoform X3 [Jatropha curcas] |
| 6 |
Hb_005218_020 |
0.1370326332 |
- |
- |
Uncharacterized protein isoform 3 [Theobroma cacao] |
| 7 |
Hb_004881_030 |
0.1379329789 |
- |
- |
PREDICTED: uncharacterized protein LOC105629532 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000586_030 |
0.1384322487 |
- |
- |
hypothetical protein OsJ_17572 [Oryza sativa Japonica Group] |
| 9 |
Hb_000237_040 |
0.1388113186 |
- |
- |
PREDICTED: V-type proton ATPase subunit a3-like [Jatropha curcas] |
| 10 |
Hb_004046_030 |
0.1393139486 |
- |
- |
PREDICTED: GPCR-type G protein 1 [Jatropha curcas] |
| 11 |
Hb_099116_010 |
0.1396176069 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000784_070 |
0.1428826387 |
- |
- |
alpha-(1,4)-fucosyltransferase, putative [Ricinus communis] |
| 13 |
Hb_004116_170 |
0.1440285751 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase dyrk1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_007383_020 |
0.1448637753 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000976_100 |
0.1468607675 |
- |
- |
eukaryotic translation elongation factor, putative [Ricinus communis] |
| 16 |
Hb_009529_040 |
0.1485454634 |
- |
- |
PREDICTED: transmembrane protein 120 homolog isoform X1 [Malus domestica] |
| 17 |
Hb_002603_050 |
0.1487560322 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP20-1-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_127552_060 |
0.1510152697 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
protein binding protein, putative [Ricinus communis] |
| 19 |
Hb_003052_030 |
0.151278721 |
- |
- |
6-phosphogluconate dehydrogenase family protein isoform 1 [Theobroma cacao] |
| 20 |
Hb_000950_010 |
0.1515067704 |
- |
- |
PREDICTED: S-formylglutathione hydrolase [Jatropha curcas] |