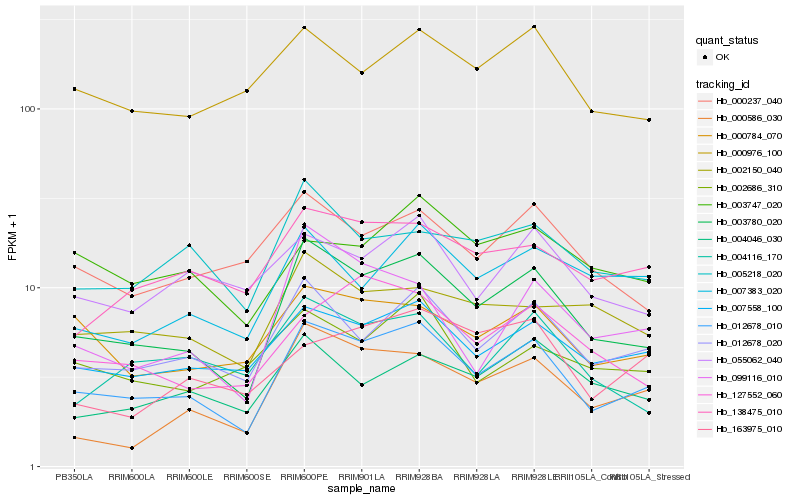
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012678_010 |
0.0 |
- |
- |
PREDICTED: outer envelope protein 61-like [Gossypium raimondii] |
| 2 |
Hb_003780_020 |
0.0582941461 |
- |
- |
PREDICTED: prolyl 4-hydroxylase subunit alpha-1 isoform X2 [Glycine max] |
| 3 |
Hb_012678_020 |
0.1034551169 |
- |
- |
PREDICTED: outer envelope protein 61 isoform X1 [Jatropha curcas] |
| 4 |
Hb_007558_100 |
0.113935354 |
- |
- |
PREDICTED: uncharacterized protein LOC105649092 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000784_070 |
0.134570536 |
- |
- |
alpha-(1,4)-fucosyltransferase, putative [Ricinus communis] |
| 6 |
Hb_138475_010 |
0.1421101027 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Populus euphratica] |
| 7 |
Hb_000586_030 |
0.1441546355 |
- |
- |
hypothetical protein OsJ_17572 [Oryza sativa Japonica Group] |
| 8 |
Hb_000976_100 |
0.1451572757 |
- |
- |
eukaryotic translation elongation factor, putative [Ricinus communis] |
| 9 |
Hb_007383_020 |
0.1453825605 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_005218_020 |
0.1457195423 |
- |
- |
Uncharacterized protein isoform 3 [Theobroma cacao] |
| 11 |
Hb_055062_040 |
0.1478031421 |
- |
- |
ceramidase, putative [Ricinus communis] |
| 12 |
Hb_127552_060 |
0.1484091767 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
protein binding protein, putative [Ricinus communis] |
| 13 |
Hb_000237_040 |
0.1492327439 |
- |
- |
PREDICTED: V-type proton ATPase subunit a3-like [Jatropha curcas] |
| 14 |
Hb_003747_020 |
0.1505552961 |
- |
- |
PREDICTED: acetyl-coenzyme A synthetase, chloroplastic/glyoxysomal isoform X1 [Jatropha curcas] |
| 15 |
Hb_004046_030 |
0.1511932758 |
- |
- |
PREDICTED: GPCR-type G protein 1 [Jatropha curcas] |
| 16 |
Hb_004116_170 |
0.1514962025 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase dyrk1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_099116_010 |
0.1517568709 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002150_040 |
0.1525589723 |
- |
- |
PREDICTED: CMP-sialic acid transporter 1 isoform X3 [Jatropha curcas] |
| 19 |
Hb_002686_310 |
0.1527027507 |
- |
- |
PREDICTED: aquaporin SIP1-1 [Jatropha curcas] |
| 20 |
Hb_163975_010 |
0.1542901208 |
- |
- |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH4 isoform X3 [Jatropha curcas] |