| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
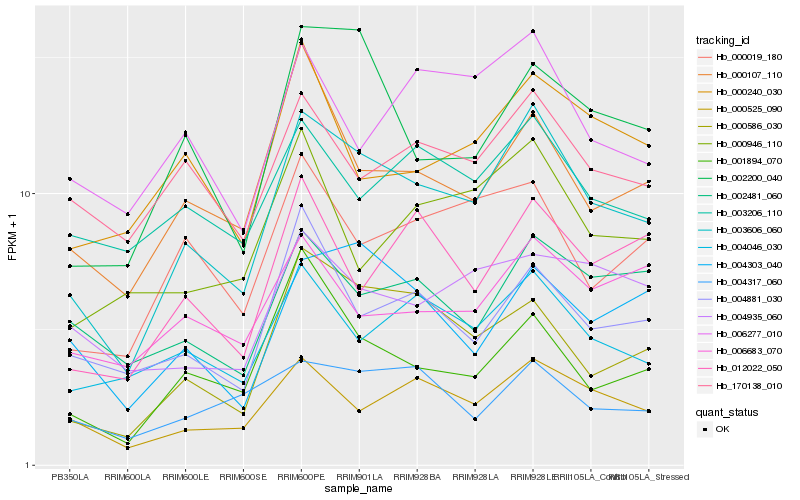
Hb_003606_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649867 [Jatropha curcas] |
| 2 |
Hb_000525_090 |
0.1247573698 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 3 |
Hb_000586_030 |
0.1253044202 |
- |
- |
hypothetical protein OsJ_17572 [Oryza sativa Japonica Group] |
| 4 |
Hb_002200_040 |
0.1288737825 |
- |
- |
PREDICTED: uncharacterized protein LOC105629143 isoform X1 [Jatropha curcas] |
| 5 |
Hb_004046_030 |
0.1320513051 |
- |
- |
PREDICTED: GPCR-type G protein 1 [Jatropha curcas] |
| 6 |
Hb_000107_110 |
0.1343166669 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 7 |
Hb_000019_180 |
0.1378867779 |
transcription factor |
TF Family: SET |
Histone-lysine N-methyltransferase family protein [Populus trichocarpa] |
| 8 |
Hb_001894_070 |
0.1401714587 |
- |
- |
PREDICTED: probable inactive poly [ADP-ribose] polymerase SRO2 isoform X1 [Jatropha curcas] |
| 9 |
Hb_004303_040 |
0.1434525567 |
- |
- |
PREDICTED: fumarate hydratase 1, mitochondrial [Jatropha curcas] |
| 10 |
Hb_004881_030 |
0.1444735674 |
- |
- |
PREDICTED: uncharacterized protein LOC105629532 isoform X1 [Jatropha curcas] |
| 11 |
Hb_170138_010 |
0.1450138935 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002481_060 |
0.1452606473 |
- |
- |
PREDICTED: uncharacterized protein LOC105640851 [Jatropha curcas] |
| 13 |
Hb_012022_050 |
0.145589886 |
- |
- |
hypothetical protein EUGRSUZ_B02804 [Eucalyptus grandis] |
| 14 |
Hb_003206_110 |
0.1461302145 |
- |
- |
Putative 1-aminocyclopropane-1-carboxylate deaminase [Gossypium arboreum] |
| 15 |
Hb_006277_010 |
0.148609903 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Gossypium raimondii] |
| 16 |
Hb_000240_030 |
0.150692387 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_006683_070 |
0.154430289 |
- |
- |
PREDICTED: uncharacterized protein LOC105631443 [Jatropha curcas] |
| 18 |
Hb_004317_060 |
0.1547731127 |
- |
- |
PREDICTED: testis-expressed sequence 10 protein isoform X2 [Jatropha curcas] |
| 19 |
Hb_000946_110 |
0.1548458534 |
- |
- |
GTP cyclohydrolase I, putative [Ricinus communis] |
| 20 |
Hb_004935_060 |
0.1555406714 |
- |
- |
Uncharacterized protein isoform 2 [Theobroma cacao] |