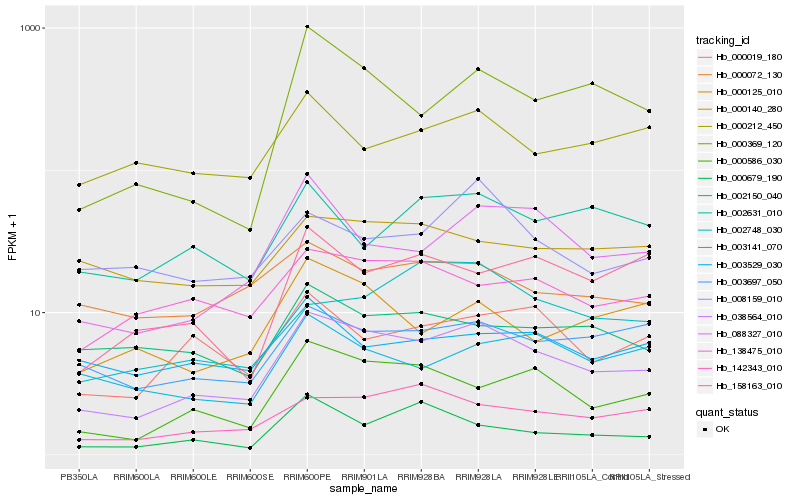
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_038564_010 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 5 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000586_030 |
0.1293704346 |
- |
- |
hypothetical protein OsJ_17572 [Oryza sativa Japonica Group] |
| 3 |
Hb_142343_010 |
0.1410792096 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 4 |
Hb_088327_010 |
0.1413235904 |
- |
- |
PREDICTED: serine hydroxymethyltransferase 7 [Jatropha curcas] |
| 5 |
Hb_003697_050 |
0.1455540031 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 13 [Jatropha curcas] |
| 6 |
Hb_000019_180 |
0.1536145262 |
transcription factor |
TF Family: SET |
Histone-lysine N-methyltransferase family protein [Populus trichocarpa] |
| 7 |
Hb_138475_010 |
0.1555114211 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Populus euphratica] |
| 8 |
Hb_000369_120 |
0.1581751913 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 3-B [Jatropha curcas] |
| 9 |
Hb_002748_030 |
0.1653556436 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 10 |
Hb_000072_130 |
0.1663563513 |
- |
- |
hypothetical protein OsJ_28567 [Oryza sativa Japonica Group] |
| 11 |
Hb_000125_010 |
0.1677507627 |
transcription factor |
TF Family: mTERF |
- |
| 12 |
Hb_000679_190 |
0.1681846474 |
- |
- |
PREDICTED: uncharacterized protein LOC105637641 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002631_010 |
0.1682110449 |
- |
- |
PREDICTED: V-type proton ATPase catalytic subunit A [Jatropha curcas] |
| 14 |
Hb_008159_010 |
0.1696449701 |
- |
- |
PREDICTED: 66 kDa stress protein [Jatropha curcas] |
| 15 |
Hb_002150_040 |
0.1698343097 |
- |
- |
PREDICTED: CMP-sialic acid transporter 1 isoform X3 [Jatropha curcas] |
| 16 |
Hb_003141_070 |
0.1700206571 |
- |
- |
PREDICTED: probable protein phosphatase 2C 51 [Jatropha curcas] |
| 17 |
Hb_158163_010 |
0.1708582015 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 18 |
Hb_000212_450 |
0.1709880476 |
- |
- |
PREDICTED: basic leucine zipper and W2 domain-containing protein 2 [Jatropha curcas] |
| 19 |
Hb_003529_030 |
0.1714286214 |
- |
- |
Non-imprinted in Prader-Willi/Angelman syndrome region protein, putative [Ricinus communis] |
| 20 |
Hb_000140_280 |
0.1721848283 |
- |
- |
hypothetical protein B456_001G157900 [Gossypium raimondii] |