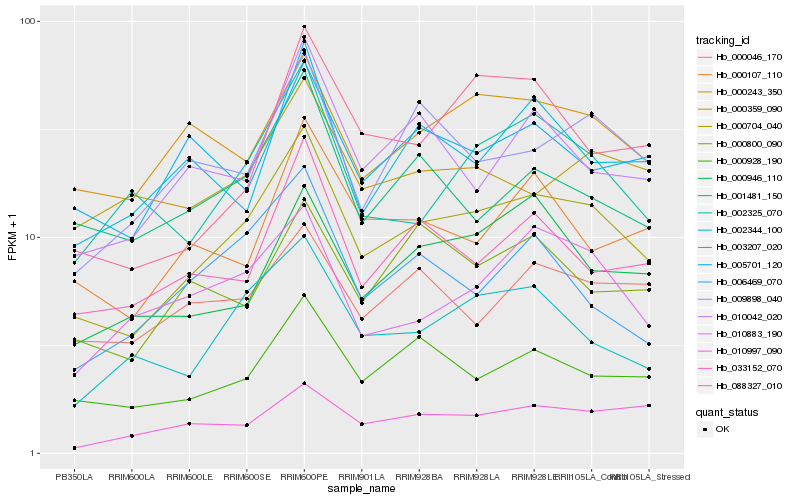
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000704_040 |
0.0 |
- |
- |
aquaporin sip2.1, putative [Ricinus communis] |
| 2 |
Hb_000928_190 |
0.11945947 |
- |
- |
PREDICTED: putative acyl-activating enzyme 19 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002344_100 |
0.1251976936 |
- |
- |
PREDICTED: cation/calcium exchanger 4-like [Jatropha curcas] |
| 4 |
Hb_033152_070 |
0.1295331507 |
- |
- |
N-acetyl-gamma-glutamyl-phosphate reductase, putative [Ricinus communis] |
| 5 |
Hb_000243_350 |
0.1303674295 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit [Tarenaya hassleriana] |
| 6 |
Hb_002325_070 |
0.1313481646 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 7 |
Hb_009898_040 |
0.1330789891 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_010042_020 |
0.133272905 |
- |
- |
UDP-n-acteylglucosamine pyrophosphorylase, putative [Ricinus communis] |
| 9 |
Hb_001481_150 |
0.1344868776 |
- |
- |
hypothetical protein POPTR_0001s15440g [Populus trichocarpa] |
| 10 |
Hb_010883_190 |
0.1375978226 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000946_110 |
0.1380428674 |
- |
- |
GTP cyclohydrolase I, putative [Ricinus communis] |
| 12 |
Hb_000107_110 |
0.1397306797 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 13 |
Hb_010997_090 |
0.142520307 |
transcription factor |
TF Family: mTERF |
hypothetical protein PRUPE_ppa019510mg, partial [Prunus persica] |
| 14 |
Hb_000800_090 |
0.1439237832 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 1 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 1 of pyruvate dehydrogenase complex, mitochondrial [Jatropha curcas] |
| 15 |
Hb_003207_020 |
0.1443730374 |
- |
- |
PREDICTED: bifunctional dTDP-4-dehydrorhamnose 3,5-epimerase/dTDP-4-dehydrorhamnose reductase [Jatropha curcas] |
| 16 |
Hb_000046_170 |
0.1444189913 |
- |
- |
PREDICTED: uncharacterized protein LOC105631841 isoform X1 [Jatropha curcas] |
| 17 |
Hb_005701_120 |
0.144451929 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit [Cucumis sativus] |
| 18 |
Hb_000359_090 |
0.1449548254 |
- |
- |
PREDICTED: persulfide dioxygenase ETHE1 homolog, mitochondrial-like [Malus domestica] |
| 19 |
Hb_006469_070 |
0.1450017341 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g22040 [Jatropha curcas] |
| 20 |
Hb_088327_010 |
0.1467549162 |
- |
- |
PREDICTED: serine hydroxymethyltransferase 7 [Jatropha curcas] |