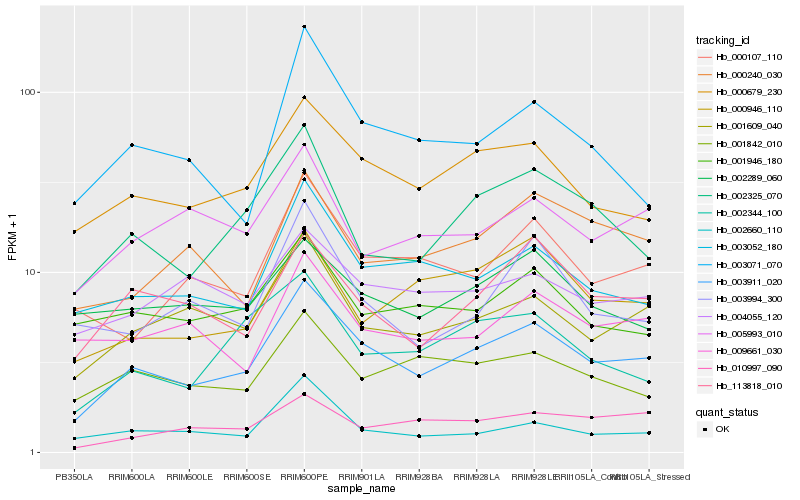
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003911_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105121332 [Populus euphratica] |
| 2 |
Hb_000679_230 |
0.1047292002 |
- |
- |
PREDICTED: serine/threonine-protein kinase PRP4 homolog [Jatropha curcas] |
| 3 |
Hb_001609_040 |
0.1144060799 |
- |
- |
hypothetical protein POPTR_0017s05650g [Populus trichocarpa] |
| 4 |
Hb_003052_180 |
0.1220011888 |
- |
- |
Diacylglycerol Cholinephosphotransferase [Ricinus communis] |
| 5 |
Hb_002325_070 |
0.1258593266 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 6 |
Hb_113818_010 |
0.1272911567 |
- |
- |
Xyloglucan galactosyltransferase KATAMARI1, putative [Ricinus communis] |
| 7 |
Hb_000107_110 |
0.1337445438 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 8 |
Hb_002344_100 |
0.1389292959 |
- |
- |
PREDICTED: cation/calcium exchanger 4-like [Jatropha curcas] |
| 9 |
Hb_001842_010 |
0.1403310651 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 10 |
Hb_002660_110 |
0.1409556448 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein CCR1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001946_180 |
0.1422110947 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_003071_070 |
0.1426353671 |
- |
- |
sucrose synthase 3 [Hevea brasiliensis] |
| 13 |
Hb_000240_030 |
0.143482324 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_005993_010 |
0.1436022062 |
- |
- |
PREDICTED: protein transport protein SEC13 homolog B [Jatropha curcas] |
| 15 |
Hb_000946_110 |
0.1441483481 |
- |
- |
GTP cyclohydrolase I, putative [Ricinus communis] |
| 16 |
Hb_010997_090 |
0.144419201 |
transcription factor |
TF Family: mTERF |
hypothetical protein PRUPE_ppa019510mg, partial [Prunus persica] |
| 17 |
Hb_004055_120 |
0.1451792288 |
- |
- |
PREDICTED: uncharacterized protein LOC105631443 [Jatropha curcas] |
| 18 |
Hb_009661_030 |
0.1482142477 |
- |
- |
PREDICTED: SPX and EXS domain-containing protein 1-like [Jatropha curcas] |
| 19 |
Hb_003994_300 |
0.1483914139 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002289_060 |
0.1487884942 |
- |
- |
conserved hypothetical protein [Ricinus communis] |