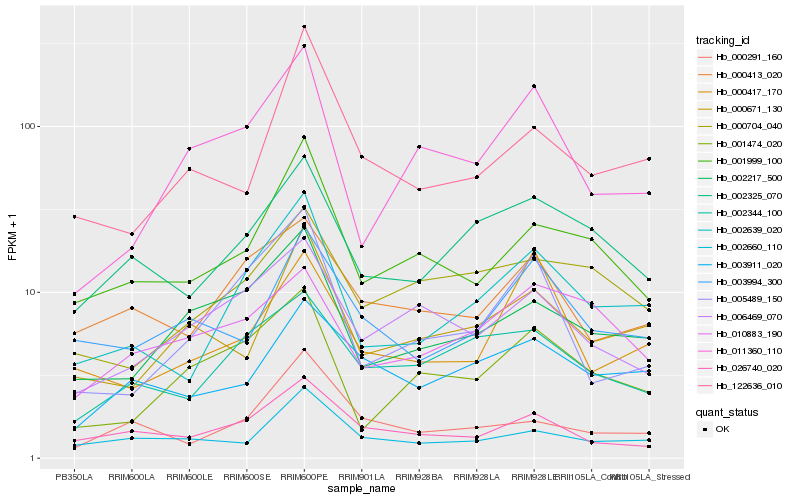
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002639_020 |
0.0 |
- |
- |
PREDICTED: probable alpha-amylase 2 [Jatropha curcas] |
| 2 |
Hb_002325_070 |
0.1369417339 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 3 |
Hb_001999_100 |
0.1585368467 |
- |
- |
PREDICTED: UDP-glycosyltransferase 71D1-like [Jatropha curcas] |
| 4 |
Hb_002217_500 |
0.1625324965 |
- |
- |
hypothetical protein JCGZ_19014 [Jatropha curcas] |
| 5 |
Hb_005489_150 |
0.1628967726 |
- |
- |
hypothetical protein JCGZ_20759 [Jatropha curcas] |
| 6 |
Hb_002344_100 |
0.1661150895 |
- |
- |
PREDICTED: cation/calcium exchanger 4-like [Jatropha curcas] |
| 7 |
Hb_001474_020 |
0.1681319356 |
- |
- |
PREDICTED: uncharacterized protein LOC105650917 [Jatropha curcas] |
| 8 |
Hb_000413_020 |
0.171040742 |
- |
- |
PREDICTED: U-box domain-containing protein 8 [Jatropha curcas] |
| 9 |
Hb_000704_040 |
0.1770889434 |
- |
- |
aquaporin sip2.1, putative [Ricinus communis] |
| 10 |
Hb_000291_160 |
0.1796178112 |
- |
- |
PREDICTED: uncharacterized protein LOC105638863 [Jatropha curcas] |
| 11 |
Hb_000671_130 |
0.1815857126 |
- |
- |
PREDICTED: CMP-sialic acid transporter 2 isoform X2 [Jatropha curcas] |
| 12 |
Hb_011360_110 |
0.1826313252 |
- |
- |
PREDICTED: UDP-glucose 6-dehydrogenase 1 [Vitis vinifera] |
| 13 |
Hb_000417_170 |
0.1846242251 |
- |
- |
PREDICTED: uncharacterized protein LOC105649689 [Jatropha curcas] |
| 14 |
Hb_026740_020 |
0.1868384062 |
desease resistance |
Gene Name: NB-ARC |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 15 |
Hb_003911_020 |
0.1887162307 |
- |
- |
PREDICTED: uncharacterized protein LOC105121332 [Populus euphratica] |
| 16 |
Hb_122636_010 |
0.1892667008 |
- |
- |
dienelactone hydrolase family protein [Populus trichocarpa] |
| 17 |
Hb_003994_300 |
0.1940299189 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_010883_190 |
0.1949910556 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002660_110 |
0.1957912521 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein CCR1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_006469_070 |
0.1971829779 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g22040 [Jatropha curcas] |