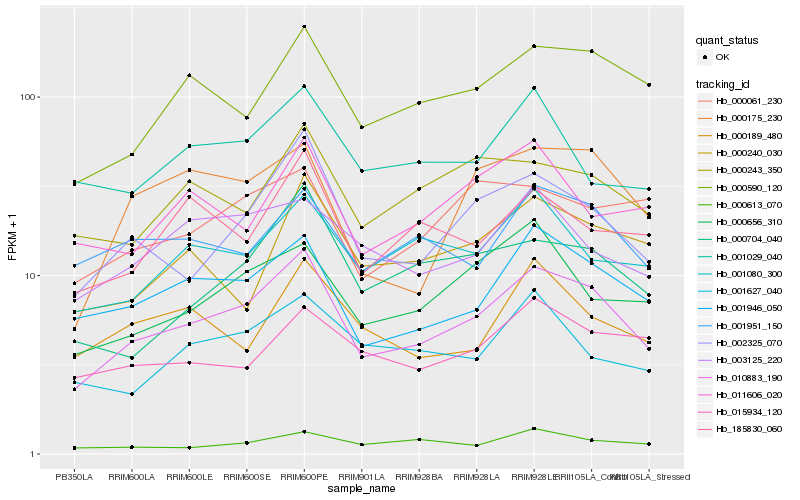
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010883_190 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002325_070 |
0.1068032267 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 3 |
Hb_001946_050 |
0.1093953641 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000590_120 |
0.1206198797 |
- |
- |
dehydroascorbate reductase, putative [Ricinus communis] |
| 5 |
Hb_000656_310 |
0.1334069421 |
- |
- |
PREDICTED: cation/calcium exchanger 4-like [Jatropha curcas] |
| 6 |
Hb_001080_300 |
0.1373276798 |
- |
- |
PREDICTED: F-box protein SKIP17-like [Jatropha curcas] |
| 7 |
Hb_000704_040 |
0.1375978226 |
- |
- |
aquaporin sip2.1, putative [Ricinus communis] |
| 8 |
Hb_000175_230 |
0.1377708638 |
- |
- |
PREDICTED: protein LURP-one-related 5 [Jatropha curcas] |
| 9 |
Hb_001951_150 |
0.1384350032 |
- |
- |
PREDICTED: uncharacterized protein LOC105649758 [Jatropha curcas] |
| 10 |
Hb_000243_350 |
0.1419994837 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit [Tarenaya hassleriana] |
| 11 |
Hb_003125_220 |
0.1423806527 |
- |
- |
PREDICTED: CBS domain-containing protein CBSCBSPB1 [Jatropha curcas] |
| 12 |
Hb_185830_060 |
0.142523422 |
- |
- |
PREDICTED: ras-related protein RABB1c [Fragaria vesca subsp. vesca] |
| 13 |
Hb_001627_040 |
0.1454941376 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 6 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000240_030 |
0.1455827711 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001029_040 |
0.1467065963 |
- |
- |
PREDICTED: uncharacterized protein LOC105641058 [Jatropha curcas] |
| 16 |
Hb_000613_070 |
0.1480864787 |
- |
- |
- |
| 17 |
Hb_011606_020 |
0.1492638798 |
- |
- |
hypothetical protein CISIN_1g023687mg [Citrus sinensis] |
| 18 |
Hb_015934_120 |
0.1497175591 |
- |
- |
PREDICTED: callose synthase 7 [Vitis vinifera] |
| 19 |
Hb_000061_230 |
0.1498421046 |
- |
- |
PREDICTED: putative glycosyltransferase 5 [Jatropha curcas] |
| 20 |
Hb_000189_480 |
0.1503367322 |
- |
- |
PREDICTED: uncharacterized protein LOC105111904 isoform X2 [Populus euphratica] |