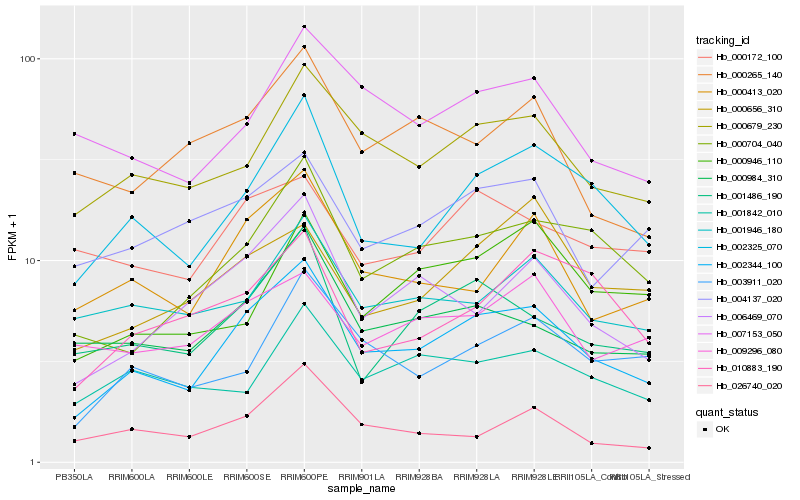
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002344_100 |
0.0 |
- |
- |
PREDICTED: cation/calcium exchanger 4-like [Jatropha curcas] |
| 2 |
Hb_002325_070 |
0.1068649053 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 3 |
Hb_000679_230 |
0.1197606992 |
- |
- |
PREDICTED: serine/threonine-protein kinase PRP4 homolog [Jatropha curcas] |
| 4 |
Hb_000704_040 |
0.1251976936 |
- |
- |
aquaporin sip2.1, putative [Ricinus communis] |
| 5 |
Hb_006469_070 |
0.1306681527 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g22040 [Jatropha curcas] |
| 6 |
Hb_000413_020 |
0.1311397421 |
- |
- |
PREDICTED: U-box domain-containing protein 8 [Jatropha curcas] |
| 7 |
Hb_000984_310 |
0.1364670221 |
- |
- |
Hydrolase, hydrolyzing O-glycosyl compounds, putative [Theobroma cacao] |
| 8 |
Hb_003911_020 |
0.1389292959 |
- |
- |
PREDICTED: uncharacterized protein LOC105121332 [Populus euphratica] |
| 9 |
Hb_001486_190 |
0.1394003615 |
- |
- |
hypothetical protein JCGZ_02702 [Jatropha curcas] |
| 10 |
Hb_007153_050 |
0.1463997981 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 8 [Jatropha curcas] |
| 11 |
Hb_001946_180 |
0.150882529 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004137_020 |
0.1520511928 |
- |
- |
PREDICTED: putative UDP-glucuronate:xylan alpha-glucuronosyltransferase 3 isoform X2 [Jatropha curcas] |
| 13 |
Hb_010883_190 |
0.1539070269 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_026740_020 |
0.1543796082 |
desease resistance |
Gene Name: NB-ARC |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 15 |
Hb_000656_310 |
0.1556889292 |
- |
- |
PREDICTED: cation/calcium exchanger 4-like [Jatropha curcas] |
| 16 |
Hb_009296_080 |
0.156468329 |
- |
- |
PREDICTED: probable apyrase 7 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000172_100 |
0.1578210446 |
- |
- |
PREDICTED: acyl-coenzyme A oxidase 2, peroxisomal [Jatropha curcas] |
| 18 |
Hb_000265_140 |
0.1579270381 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 12 [Jatropha curcas] |
| 19 |
Hb_001842_010 |
0.158259154 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 20 |
Hb_000946_110 |
0.1604138527 |
- |
- |
GTP cyclohydrolase I, putative [Ricinus communis] |